Genomic and chromosomal change as motor of evolution
The blueprint of all life is the DNA, which in its entirety is called the genome. The genome is constantly changing due to a multitude of factors. Some changes can become established and are passed on to descendants. Some changes are so strong that even short evolutionary periods of time are sufficient to produce new species and to generate biodiversity. The research of the Chair of Cell and Molecular Biology of Plants focuses on the mechanisms that reorganize and comprehensively alter genomes: genome duplications, the activation of mobile DNA elements and the restructuring of entire chromosomes.
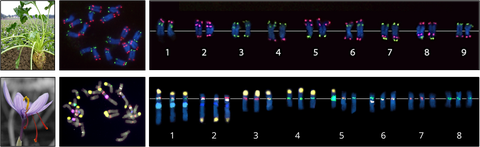
The figure shows how chromosomal markers were obtained from high-throughput sequencing data for the unique assignment of chromosomes. Top: The 18 chromosomes of the sugar beet can be clearly divided into nine pairs and distinguished by a chromosomal barcode. As a molecular basis 54,720 oligonucleotides were anchored to the chromosomes. Bottom: 92 chromosomal markers allow clear identification of the individual chromosomes of the triple genome of saffron crocus.
We work at the interface between molecular biology, cytogenetics and bioinformatics. We are also involved in the sequencing, assembly and annotation of plant genomes, e.g. of crops such as sugar beet and saffron crocus. Since the number, size and morphology of chromosomes varies extremely between plant species, we have established high-resolution methods for chromosome analysis. Fluorescent in situ hybridisation (FISH) is the only technique that allows to localise the location and distribution of each DNA sequence on the chromosomes. The insights into chromosome structure and evolution gained in this way complement our bioinformatics approaches and thus provide us with a unique stand-alone position: Since we can cytogenetically anchor our bioinformatics data onto the chromosomes, we make a considerable contribution to the generation of reference genome sequences of the highest quality.
Selected publications
Heitkam T., Weber B., Walter I., Liedtke S., Ost C. and Schmidt T. (2020): Satellite DNA landscapes after allotetraploidisation of quinoa (Chenopodium quinoa) reveal unique A and B subgenomes. The Plant Journal, doi: 10.1111/tpj.14705 read article
Read the corresponding preprint (open access): bioRxiv, doi: 10.1101/774828
Schmidt T., Heitkam T., Liedtke S., Schubert V. and Menzel G. (2019): Adding color to a century-old enigma: Multi-color chromosome identification unravels the autotriploid nature of saffron (Crocus sativus) as a hybrid of wild Crocus cartwrightianus cytotypes. New Phytologist, 222(4): 1965-1980 read article
Rodríguez del Río Á., Minoche A., Zwickl N., Friedrich A., Liedtke S., Schmidt T., Himmelbauer H. and Dohm J. (2019): Genomes of the wild beets Beta patula and Beta vulgaris ssp. maritima. The Plant Journal 99(6): 1242-1253 read article
Dohm JC, Minoche AE, Holtgräwe D, Capella-Gutiérrez S, Zakrzewski F, Tafer H, Rupp O, Sörensen TR, Stracke R, Reinhardt R, Goesmann A, Kraft T, Schulz B, Stadler PF, Schmidt T, Gabaldón T, Lehrach H, Weisshaar B, Himmelbauer H, (2014): The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 505: 546–549 read article
