Dec 29, 2020
New article on the ecology of plasmid-coded antibiotic resistance

Entities, state variables and scales affecting the ecology of plasmid-coded antibiotic resistance (from Zwanzig 2020, https://doi.org/10.1016/j.csbj.2020.12.027).
A number of entities, state variables, and scales are relevant to the spread of plasmid-encoded antibiotic resistance in aquatic environments. This review article summarizes relevant plasmid properties, ecological factors, and evolutionary processes and presents an outlook on appropriate modeling approaches.Plasmids vary in size and can provide antibiotic resistance genes (ARG) or phage resistance to their host. This can help cells to prevent growth inhibition or cell death in consequence of the presence of some antibiotic. If plasmids carry transfer genes (tra), they can perform conjugation, which transfers a copy of the plasmid from the host to a nearby recipient cell. Bacteria can be either solitary or part of a microcolony or biofilm. Those that detach from the biofilm may be transported to another location where they can exchange genes with other subpopulations or members of the microbial community.
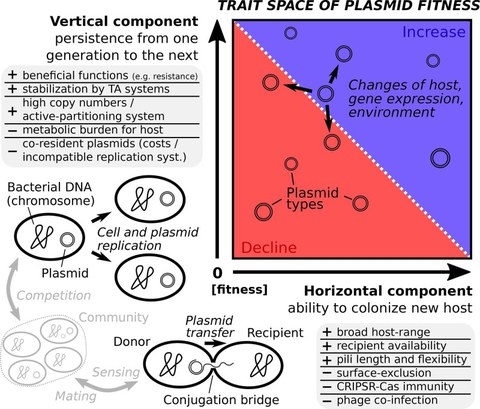
Trait space of plasmid fitness (from Zwanzig 2020, https://doi.org/10.1016/j.csbj.2020.12.027).
A plasmids frequency in a local community declines if vertical transmission fitness is low, e.g. due to high rates of missegregation and plasmid costs, and if this cannot be compensated by horizontal transmission, e.g. conjugation. Both vertical and horizontal transmission fitness are not constant for a specific plasmid, but can be host-specific and change over time due to evolutionary modifications and spatio-temporal variations of environmental factors.
Take a look at the full Open Access article by clicking on the link below:
Zwanzig, M. (2021): The ecology of plasmid-coded antibiotic resistance: a basic framework for experimental research and modeling. Computational and Structural Biotechnology Journal, 19, 586 - 599.
