Research of the Plant Genomics Group
Although plant genomes contain all information necessary for building whole organisms, we are still far from understanding this molecular building plan. Nevertheless, being able to read out any genome offers ways towards protecting the existing plant biodiversity, towards predicting how any organism may react to changing environmental conditions or biotic and abiotic stresses, and towards guiding plant breeding. Especially at the current times, during which we start to feel the impact of climate change, the ability to produce maximally informed decisions towards one or several plant traits will strongly benefit plant and ecosystem resilience.
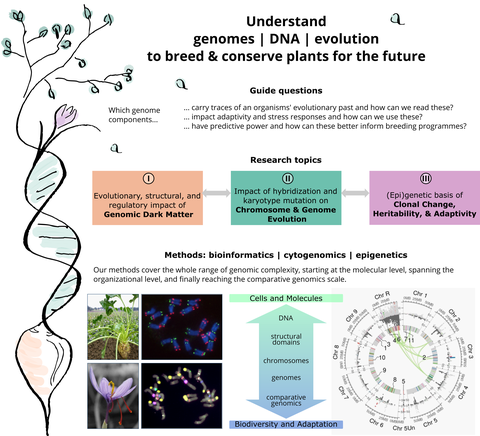
This can be summed up in our central mission: Understanding our plants' building plan and its evolution to breed and conserve plants for the future.
Specifically, we are interested in genome-, epigenome, and chromosome-wide changes, their impact on adaptivity, and their control across different time scales – ranging from immediate stress responses over the life span of an individual up to domestication/ evolutionary time scales. For this, our research is organized along three pillars:
|
Research in Pillar 1: Genomic Dark Matter
Two of the fastest routes towards genomic change are the activation and restructuration of repetitive DNAs, namely transposable elements and satellite DNAs. We recognize repetitive DNA as raw material for genome evolution and regulatory innovation:
- Repetitive DNAs are not only the size-determining fraction of plant genomes, they provide the genome’s structural backbone, constitute centromeres, and delimit chromosome ends. Hence, knowledge of the repeat fraction can be directly used to gain insights into genome and chromosome evolution of organisms, e.g. for conifers.
- If active, repetitive DNAs can rapidly speed up evolutionary processes. Their distribution and abundance across genomes can be used to clarify the evolutionary history of the species, as investigated for saffron, quinoa, and poplars.
- After the genomic integration of a potentially disease-causing agent, such as a virus or a transposable element, this insertion can be quickly fragmented, silenced, or densely packed. Hence, all organisms operate at a tight balance between genomic change and control.
- If situated close to genes, repetitive DNAs can supply promoter regions, splice sites, transcription starts and stops, as shown across a wide range of plant genomes. Thus, repetitive DNAs are the molecular agents of choice to enable fast genomic change, adaptation, and ultimately speciation and biodiversity.
Research in Pillar 2: Chromosome & Genome Evolution
In many cases, knowledge of the repeat fraction can be directly used to gain insights into plant genome and chromosome evolution. The fast sequence turn-over of repeat components makes them immensely useful to determine the evolutionary history of organisms, e.g. to trace the genomic origin of introgressions or of the parental species of polyploids as exemplified by the following two case studies:
- The genetic origin of saffron crocus: We have applied this to determine the genetic origin of the triploid saffron crocus (Crocus sativus), source of saffron, the most expensive spice of the world. Due to its triploidy, saffron crocus is sterile, can only be propagated vegetatively, and is inaccessible for breeding. Skim sequencing allowed identifying the major repeats and converting them to cytogenetic landmarks. Using nearly 100 landmark probes for fluorescent in situ hybridization, it was possible to determine that C. sativus resulted from hybridization of two cytotypes of Crocus cartwrightianus.
- The origin of quinoa’s tetraploidy: Similarly, using quinoa (Chenopodium quinoa) as a tetraploid model with undefined origin, we have traced the distribution of tandem repeats from putative diploid parents to allotetraploid quinoa. This allowed insights into species-specific tandem repeat behavior, the different impact on the two subgenomes, shedding light on the evolution of the species quinoa.
Research in Pillar 3: Epigenetics of Clonality, Heritability, and Adaptivity
The repetitive genome, especially transposable elements, is tightly controlled by a complex interplay of epigenetic mechanisms, including DNA methylation maintenance. We investigate the impacts of disturbed DNA methylation as often caused by environmental or biotic stresses, employing suitable natural systems and methylation-deficient mutants. This will give insights into the molecular basis of crop adaptability, and into a genome’s ability to control and react to change, as exemplified by one of our current projects:
- Identical genomes but different properties – saffron as a model for epigenetics of quality traits and environmental adaptation: We investigate how clonal, genetically identical plants can have different phenotypes depending on where they grow. We zoom in on the epigenetics of different saffron accessions to identify the molecular basis of different phenotypes for one genome.
Funded projects
-
Identical genomes but different properties – saffron as a model for epigenetics of quality traits and environmental adaptation (funded by DFG) [ongoing]
-
EpicBeet – Stability and heritability of DNA methylation patterns in sugar beet: impact on phenotypic plasticity, influence on TE activity and potential for beet breeding (funding by BMBF) [ongoing]
-
LarchForFlexibility – Provision of larch reproductive material with high quality and diversity to increase silvicultural flexibility (funding by BMEL; FNR) [to be started]
- Cytogenetics of mango germplasm (we are glad to host Dr. Sultana during her research stay that is funded by the Alexander von Humboldt foundation) [finished]
- SINEs4Conifers – Genotypisierung der Fichte für die Qualitätskontrolle und Identitätssicherung von Forstvermehrungsgut (funding by BMEL; FNR) [finished]
- MASEPA – Markergestützte Selektion in der Züchtung von Kartoffeln mit Resistenz gegen Globodera pallida (BMBF; KMU-innovativ) [finished]