DNA Transposons
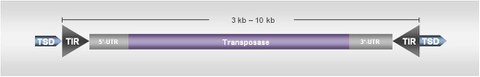
In contrast to retrotransposons, class II DNA transposons mostly code for a single gene product designated as transposase. The transposase gene is flanked by 5’- and 3’-untranslated regions and terminal inverted repeats (TIRs). DNA transposons are divided in the four classes of "cut and paste", "self-synthesizing", "replicative", and "rolling-circle", thus describing their mechanisms of mobilization.
In a cut and paste mobilization, the transposon is enzymatically cut from the insertion site by the transposase. The integration into a new genomic position results in the doubling of a short nucleotide motif of the insertion site, designated as target site duplication (TSD). TIRs and TSDs are specific within the five major cut and paste transposon superfamilies Mariner, CACTA, PIF/Harbinger, Mutator and hAT. Each of these superfamilies also contain MITEs (Miniature Inverted-Repeat Transposable Elements), representing non-autonomous derivatives lacking the transposase coding capacity.
Based on unique amino acid motifs of the transposase, we have isolated several members of different transposon superfamilies from a sugar beet BAC-library. Beside Mariner and hAT cut and paste transposons, we have identified Helitrons, which are mobilized by a rolling circle mechanism mediated by at least two Helitron-encoded enzymes.
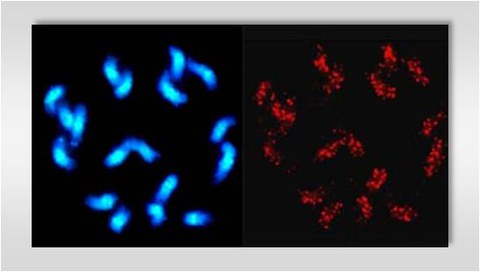
Southern hybridizations reveal a dispersed genomic organization of Mariner and hAT transposons in species of the genus Beta. Fluorescent in situ hybridization (FISH) on B. vulgaris metaphase chromosomes indicate the specific chromosomal distributions of the autonomous members and MITEs of both transposon families.
PCR analyses show the transposition of the Mariner derived MITE VulMITE I in distinct sugar beet cultivars. This indicates a recent mobilization of VulMITE I during domestication of sugar beet, which started in the late 18th century.
Related publications
- Menzel et al. (2014): The diversification and activity of hAT transposons in Musa genomes. Chromosome Research, 22(4):559-571 read article
- Menzel et al. (2012): Survey of sugar beet (Beta vulgaris) hAT transposons and MITE-like hATpin derivatives. Plant Molecular Biology 78(4-5):393-405 read article
- Menzel et al. (2006): Mobilization and evolutionary history of minitiature-inverted-repeat transposable elements (MITEs) in Beta vulgaris L. Chromosome Res. 14:831-844 read article