News
MAR 2021 // Abdullah El-nagish will join the lab as a new PhD student to work with us on saffron crocus
Abdullah El-nagish has received a scholarship from the Egyptian Ministry of Higher Education and arrived to join the work on saffron as a PhD student. He is coming from Sohag University, Egypt with a Master’s degree in Plant Biotechnology and Physiology and will join the work on saffron to determine the genetic and epigenetic foundations of saffron variations.
More information about Abdullah can be found here:
https://www.researchgate.net/profile/Abdullah-El-Nagish
https://scholar.google.com.eg/citations?user=o7BKgRcAAAAJ&hl=en
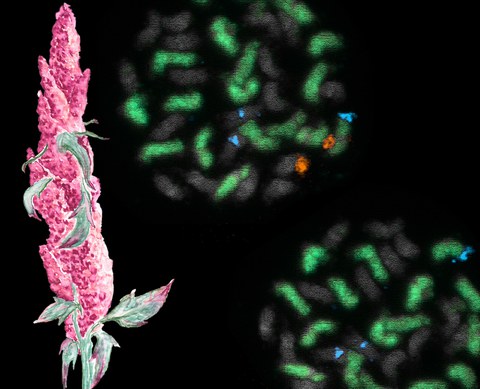
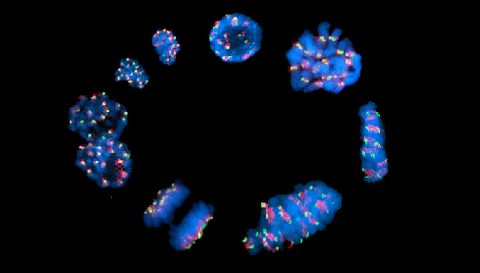
DEC 2020 // Contribution to the 4th Chromosome Calendar
Happy to have contributed to the 4th Chromosome Calender, providing the image for July! The calendar is a cooperative effort led by the IPK Gatersleben. A pdf of the calendar can be downloaded and printed to decorate your office wall. If you want to know more about the chromosome image, you will find this information in our preprint.
OCT 2020 // Dr. Nusrat Sultana has received an Alexander von Humboldt fellowship to work on mangos from Bangladesh
We are looking forward to work with Dr. Nusrat Sultana from the Jagannath University in Bangladesh on mango genomics and cytogenetics. Bangladesh is one of the presumed centers of domestication for mango with many local wild species and cultivated varieties. We will provide information on the genomic diversity of Bangladesh's mangos to support local strategies for germplasm conservation.
More information on Dr. Sultana and her home university
More information on the fellowship
SEP 2020 // New Paper: Cassandra retrotransposons are a prime target for genomic rearrangements in beet
We are happy to announce that our paper on Cassandra retrotransposons in beet and related Amaranthaceae has just been published by Annals of Botany. Genome analyses showed homology-mediated Cassandra restructuring and suggested that reshuffling and recombination has strongly affected this small non-autonomous mobile element and the evolution of the beet genome.
Maiwald et al. (2020): The Cassandra retrotransposon landscape in sugar beet (Beta vulgaris) and the Amaranthaceae: Recombination and re-shuffling lead to a high structural variability. Annals of Botany, doi: 10.1093/aob/mcaa176 read article (free access link)
JAN 2020 // New Paper: Revealing unique A and B subgenomes in quinoa
Quinoa originated by hybridization of an unknown female American Chenopodium diploid (AA genome) with an unknown male Old World diploid species (BB genome). Whereas the satDNA distributions support C. suecicum as possible parental species, we were able to exclude C. pallidicaule as progenitor due to unique repeat profiles.
Heitkam et al. (2020) Satellite DNA landscapes after allotetraploidization of quinoa (Chenopodium quinoa) reveal unique A and B subgenomes. The Plant Journal, 103: 32-52 read article
Jan 2020 // DFG funding to continue our work on saffron (epi)genomics
"Identical genomes but different properties – saffron as a model for epigenetics of quality traits and environmental adaptation": Our collaborative effort with the IPK Gatersleben and the FZ Jülich to sequence the saffron crocus has been funded by the DFG. Comparing accessions world-wide, saffron crocus is nearly genetically identical. Nevertheless, this special crop is adapted to the environments of its cultivation, with differences in its quality traits. We want to test if epigenetic changes are the cause.
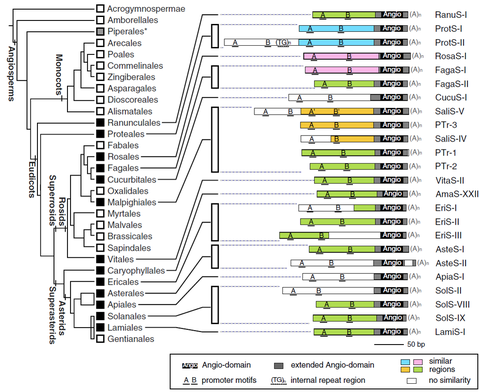
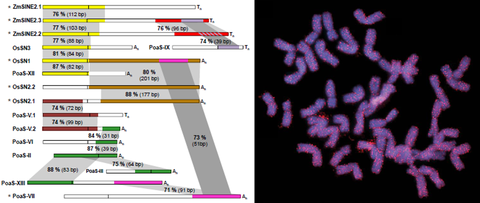
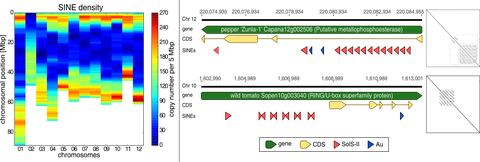
OCT 2019 // New Paper: The Angio-SINE superfamily in higher plants
Short interspersed nuclear elements (SINEs) are non-coding retrotransposons that contain only few short conserved structural motifs. SINEs emerge and vanish during evolution, and often diversify into numerous families and subfamilies that are usually specific for only a limited number of species. In contrast, at the 3' end of multiple plant SINEs we detected the highly conserved ‘Angio-domain’.
Seibt et al. (2019) The conserved 3′ Angio‐domain defines a superfamily of short interspersed nuclear elements (SINEs) in higher plants. The Plant Journal, 101: 681–699 read article
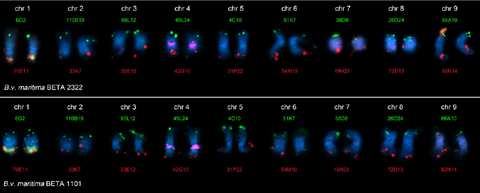
JUL 2019 // New Paper: New genome assemblies of two wild beet species
We present draft genome assemblies of Beta patula, a critically endangered wild beet endemic to the Madeira archipelago, and of the closely related Beta vulgaris ssp. maritima (sea beet). Large syntenic regions were identified, and a display tool for automatic genome-wide synteny image generation was developed.
del Rio et al. (2019) Genomes of the wild beets Beta patula and Beta vulgaris ssp.
maritima. The Plant Journal, 99: 1242–1253 read article
MAY 2019 // Saffron article highlighted on New Phytologist's blog
We are happy to share that our latest article "Multi-color chromosome identification unravels the autotriploid nature of saffron" made the cover of the current
New Phytologist issue and was highlighted on New Phytologist's blog.
read more: New Phytologist's blog
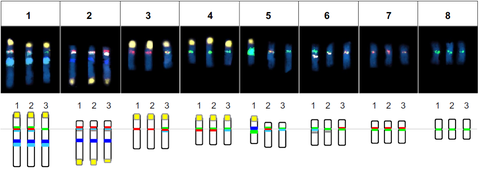
JAN 2019 // New Paper: Multi‐color chromosome identification unravels the autotriploid nature of saffron
We performed a survey sequencing of the saffron genome and selected cytogenetic landmarksequences consisting of major tandem repeats, which we used as probes in comparativemulticolor fluorescent in situ hybridization (FISH). We demonstrate
that Crocus sativus is an autotriploid hybrid derived from heterogeneous Crocus cartwrightianus cytotypes.
Schmidt et al. (2019) Adding color to a century‐old enigma: multi‐color chromosome identification unravels the autotriploid nature of saffron (Crocus sativus ) as a hybrid of wild Crocus cartwrightianus cytotypes. New Phytologist, 222: 1965–1980 read article
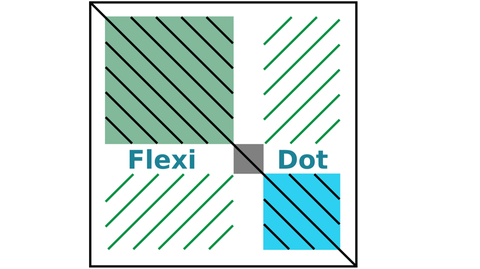
MAY 2018 // New Paper: FlexiDot - a new tool for visual sequence analyses
FlexiDot is a cross-platform dotplot suite generating high quality self, pairwise and
all-against-all visualizations. Combined with collage-like outputs, FlexiDot supports simultaneous visual screening of large sequence sets, enabling dotplot use for routine analyses. access on GitHub
Seibt et al. (2018) FlexiDot: highly customizable, ambiguity-aware dotplots for visual sequence analyses. Bioinformatics, 34: 3575–3577 read article
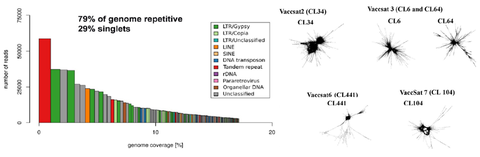
OCT 2017 // New Paper: Genome differences between the northern highbush blueberry and the American cranberry
In this study, the major genomic differences between Vaccinium corymbosum (northern highbush blueberry) and Vaccinium macrocarpon (American cranberry) were discovered through comparative analysis of repetitive sequences using the RepeatExplorer software. V. macrocarpon contains remarkably higher amounts of LTR and non-LTR retrotransposon, whereas the percentage of satellite repeats was significantly higher in V. corymbosum.
Sultana et al. (2017) Comparative Analysis of Repetitive Sequences Reveals Genome Differences between Two Common Cultivated Vaccinium Species (V. corymbosum and V. macrocarpon). Journal of Molecular Biology and Biotechnology, 1: 07-15 read article
AUG 2017 // New Paper: Diversity of short interspersednuclear element (SINE) families in grasses
We comparatively analyzed the structural features, distribution, evolutionary relation and abundance of 32 SINE families and subfamilies within grasses and found that multimerization of related and unrelated SINE copies is an important evolutionary mechanism of SINE formation.
Kögler et al. (2017) Evolutionary modes of emergence of short interspersed nuclear element (SINE) families in grasses. Plant Journal, 92: 676– 695 read article
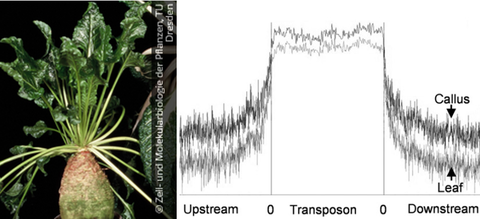
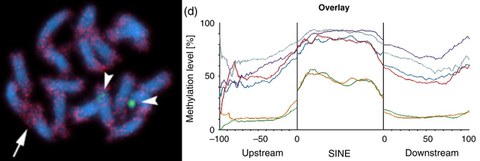
MAR 2017 // New Paper: Unraveling the methylation state of repetitive DNAs in beet
Genome-wide cytosine methylation in the sugar beet genome was studied in leaves and leafderived callus with a focus on repetitive sequences and compared with gene methylation. Comparison of genome-wide DNA methylation between sugar beet leaves and callus revealed a differential methylation upon tissue culture.
Zakrzewski et al. (2017) DNA methylation of retrotransposons, DNA transposons and genes in sugar beet (Beta vulgaris L.). Plant Journal, 90: 1156–1175 read article
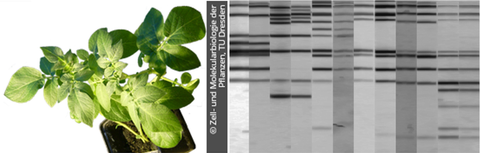
JAN 2017 // New Paper: Diversity studies in potato species using the ISAP marker system
Here, we describe the experimental application of the retrotransposon-based Inter-SINE amplified polymorphism (ISAP) marker system for supporting the management of potato collections. Compared to microsatellites, ISAPs were similarly efficient in monitoring genetic diversity and detecting duplicated accessions. Generally, ISAPs are a very valuable tool in the maintenance of clonally maintained potato collections and for the verification of the identity of accessions.
Diekmann et al. (2017) Diversity studies in genetic resources of Solanum spp. (section Petota) by comparative application of ISAP markers. Genet Resour Crop Evol, 64: 1937–1953 read article
MAY 2016 // New Paper: ChIP-Seq unveils heterochromatin sequence composition
CenH3-chromatin and H3K9me2-heterochromatin are only poorly characterized. Here we identify the associated repetitive genome portions by CenH3 and H3K9me2 ChIP-seq. Our experiments complement the sugar beet reference sequence and provide insight into the repeat composition of heterochromatic regions.
Kowar et al. (2016) Repeat composition of CenH3- chromatin and H3K9me2-marked heterochromatin in sugar beet (Beta vulgaris). BMC Plant Biology, 16:120 read article
MAR 2016 // New Paper: Solanaceae SINEs and their impact on genes and genome
Comparative genomics of short interspersed nuclear elements (SINEs) are rare. Here we document species- and family-specific amplifications of SINEs in the Solanaceae, showing that they are unevenly dispersed and strongly enriched in gene-rich chromosomal regions, particularly in intron and untranslated regions, where they likely contribute to gene and genome evolution.
Seibt et al. (2016) Short interspersed nuclear elements (SINEs) are abundant in Solanaceae and have a family-specific impact on gene structure and genome organization. The Plant Journal, doi 10.1111/tpj.13170 read article
JAN 2016 // New Paper: Beet SINEs in The Plant Journal
Although important players in gene regulation, plant SINEs are not well understood and largely underrepresented in current genome annotations. Here, we identify and characterize the SINE population in sugar beet and related species, comprising 34,806 SINEs, including 19,549 full length copies. This database of plant SINEs allowed us to perform a unprecedented comparative analysis to understand the diversity, evolution, chromosomal organization and cytosine methylation of SINEs.
Schwichtenberg et al. (2016) Diversification, evolution and methylation of Short Interspersed Nuclear Element families in sugar beet and related Amaranthaceae species. The Plant Journal, 85:229-244 read article
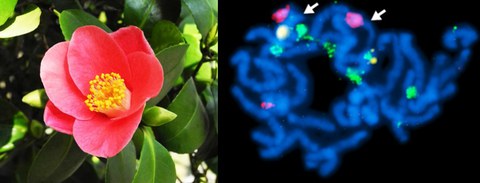
NOV 2015 // New Paper: Satellite DNA is a major component of the Pillnitz camellia's genome
Together with our colleagues from the TU Dresden Chair of Botany, the TU Dresden Botanical Garden and the Schlösser, Burgen und Gärten Sachsen gGmbH, we analyse sequence and composition of the Pillnitz camellia's genome. Here, we publish a first view into the Pillnitz camellia genome: Using next-generation sequencing and read clustering, we classified the main repeat families and estimated a camellia repeat fraction of 73 %. Focussing on tandem repeats, we characterize its satellite DNAs and the 5S ribosomal DNA by bioinformatics, fluorescent in situ and Southern hybridization. This work provides an insight into the C. japonica chromosome organization and significantly expands the Camellia genomic knowledge, also with respect to the tea plant Camellia sinensis.
Heitkam et al. (2015): Next-generation sequencing reveals differentially amplified tandem repeats as a major genome component of Northern Europe's oldest Camellia japonica. Chromosome Research, 23(4):791-806 read article
in Focus // Nature Article: Sugar beet reference genome sequence
We are happy to have contributed to the published genome sequence of sugar beet published in Nature. You can find the genome sequence and information on all partners on the Beta vulgaris Resource.
Dohm et al. (2014) : The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 505:546–549. read article