LTR retrotransposons
LTR retrotransposons represent the most widespread class of TEs in plants. Their proliferation by reverse transcription of RNA intermediates can rapidly increase the copy number of elements and can thereby greatly increase plant genome size. However, their presence can also counteract genome expansion through illegitimate recombination. They are flanked by long terminal repeats (LTRs), which drive reverse transcription using promoter acting motifs. The proteins essential for the reverse transcription and integration of a new LTR retrotransposon copy are encoded as a gag-pol-polyprotein in one or two open reading frames (ORFs). Based on the order of encoded gene products the LTR retrotransposons can be further subclassified into the Ty3-gypsy family (Metaviridae), the Ty1-copia family (Pseudoviridae), and the BEL-Pao family, where the latter one described only in metazoan genomes to date.
Because of the chromosomal dynamics of LTR retrotransposons their influence on structure and genome evolution is focussed in our research. Therefore, the structural and functional study of LTR retrotransposons as an abundant component of genomes in the genus Beta and Patellifolia in particular with restricted contribution (centromeric retrotransposons) or additional features (env-like ORF) are on high interest.
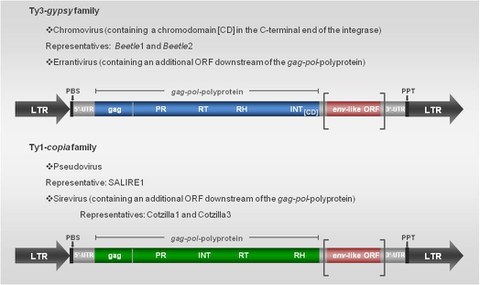
Structure of two retrotransposon types: The blue, green and red boxes indicate translated regions. LTR: long terminal repeat, gag: gag-like protein, PR: protease, RT: reverse transcriptase, RN: RNase H, INT: integrase, CD: chromodomain, PBS: primer binding site, PPT: polypurine tract, UTR: untranslated region, ORF: open reading frame.
Their transcriptional activity combined with high sequence conservation within each family and the exclusively localization in centromeric heterochromatin suggest, that these transposable elements may play an important role in the centromeres of P. procumbens chromosomes.
A non-random genomic distribution of LTR retrotransposons and an accumulation in specific chromosomal regions, especially the clustering in silent heterochromatin is apparent. A role of the integrase for different patterns of integration, particularly the additional presence of a chromodomain (CD) at the C-terminal end may play a potential role for the target specificity of LTR retrotransposons in the host genome. Ty3-gypsy retrotransposons, which posses a chromodomain are generally referred to as chromoviruses. In plants four chromoviral clades (CRM, Tekay, Galadriel, Reina) are present. Interestingly, the chromodomain of centromeric plant Ty3-gypsy retrotransposons (CRs) is substantially divergent from the overall.
Furthermore, in plant genomes LTR retrotransposons could be identified that habour an additional ORF downstream of the gag-pol polyprotein. These LTR retrotransposons are related to retroviruses and the additional ORF contains domains reminiscent to envelope (env)-genes of retroviruses. Env-like retrotransposons are present in the Ty3-gypsy family (Errantivirus) and Ty1-copia family (Sirevirus), respectively. Members of each family were characterized in Beta species after screening of BAC- and cot1 DNA libraries.
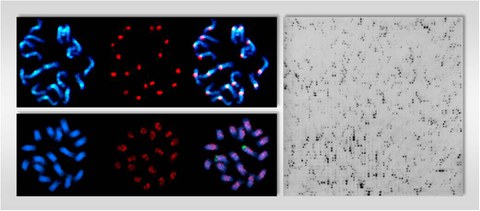
Physical mapping of LTR retrotransposons (red signals) by fluorescent in situ hybridization (FISH) on metaphase chromosomes of Beta species, showing a restricted localization in centromeric regions (A) or dispersed distribution along the chromosomes (B). The DAPI-stained DNA (blue fluorescence) in panel A and B shows the morphology of the chromosomes. C) Southern hybridization of a high-density BAC filter using a DNA probe specific for the reverse transcriptase region of an env-like retrotransposon.
Related publications
- Weber et al. (2013): Highly diverse chromoviruses of Beta vulgaris are classified by chromodomains and chromosomal integration. Mobile DNA, 4:8. read article
- Wollrab et al. (2012): Evolutionary reshuffling in the Errantivirus lineage Elbe within the Beta vulgaris genome. Plant Journal, 72:636-651 read article
- Weber et al. (2010) :The Ty1-copia families SALIRE and Cotzilla populating the Beta vulgaris genome show remarkable differences in abundance, chromosomal distribution, and age. Chromosome Research, 18:247-263. read article
- Weber and Schmidt (2009): Nested Ty3-gypsy retrotransposons of a single Beta procumbens centromere contain a putative chromodomain. Chromosome Research, 17:379-396 read article
