Publikationen
Bitte kontaktieren Sie uns, falls sie an einer Kopie unserer Publikationen interessiert sind.
2025
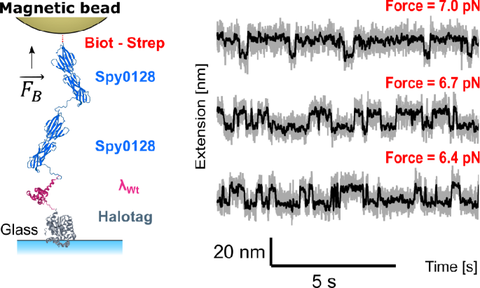
Magnetic tweezers to capture the fast-folding λ6-85 in slow motion
CA. Qunitana-Cataño, A. Mukhortava, MA. Boukhet, A. Hartmann, KL. Schuhmann, M. Schlierf
Communications Physics
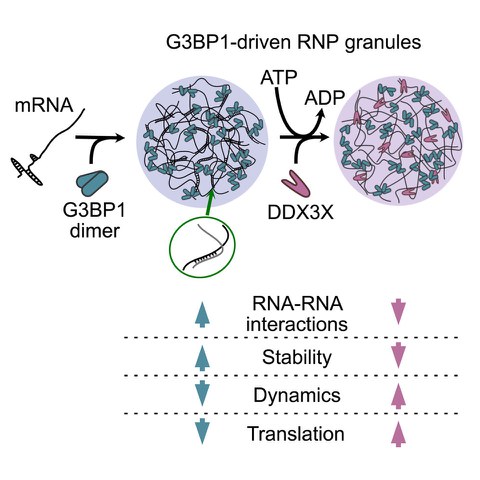
G3BP-driven RNP granules promote inhibitory RNA-RNA interactions resolved by DDX3X to regulate mRNA translatability
IREA. Trussina, A. Hartmann, C. Desroches Altamirano, J. Natarajan, CM. Fischer, M. Aleksejczuk, H. Ausserwöger, TPJ. Knowles, M. Schlierf, TM. Franzmann, S. Alberti
Molecular Cell
2024
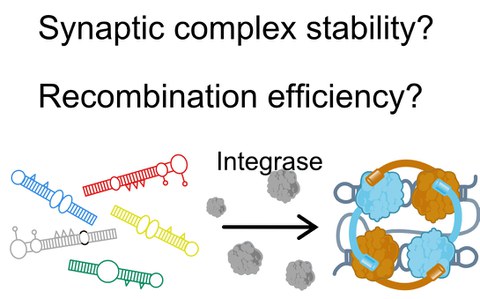
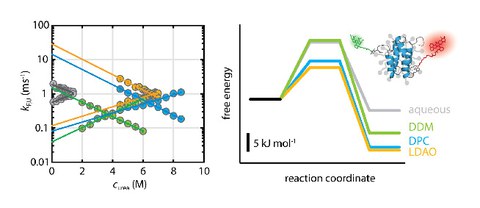
The recombination efficiency of the bacterial integron depends on the mechanical stability of the synaptic complex
E. Vorobevskaia, C. Loot, D. Mazel, M. Schlierf$
Science Advances
BioRxiv
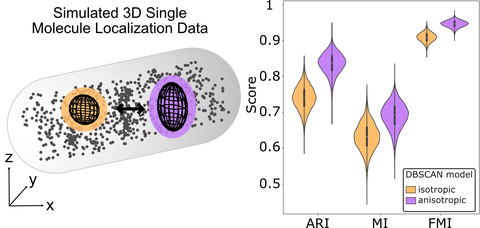
Anisotropic DBSCAN for 3D SMLM Data Clustering
P. Lörzing*, P. Schake*, M. Schlierf
JPC B
2023
Thermophoretic Analysis of Biomolecules across the Nanoscales in Self-Assembled Polymeric Matrices
P. Liu, N. Schumann, F. Abele, F. Ren, M. Hanke, Y. Xin, A. Hartmann, M. Schlierf, A. Keller, W. Lin$, Y. Zhang$
ACS Applied Nano Materials
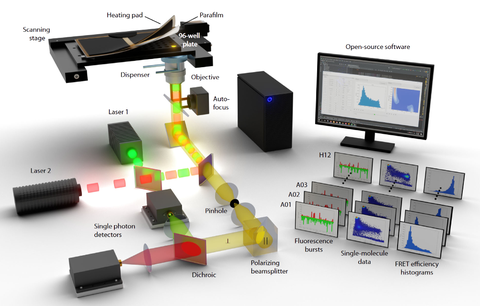
An automated single-molecule FRET platform for high-content, multiwell plate screening of biomolecular conformations and dynamics
A. Hartmann$, K. Sreenivasa*, M. Schenkel*, N. Chamachi*, P. Schake*, G. Krainer, M. Schlierf$
Nature Communications
Software on github
Twofold mechanosensitivity ensures actin cortex reinforcement upon peaks inmechanical tension
V. Ruffine, A. Hartmann, M. Schlierf, E. Fischer-Friedrich
Advanced Physics Research
Arxiv
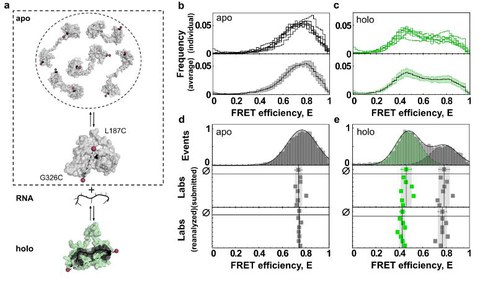
Reliability and accuracy of single-molecule FRET studies for characterization of structural dynamics and distances in proteins
G. Agam, C. Gebhardt, M. Popava, et int, N. Chamachi, A. Hartmann, M. Schlierf, et al.
Nature Methods
SI
Farewell to single-well: An automated single-molecule FRET platform for high-content, multiwell plate screening of biomolecular conformations and dynamics
A. Hartmann$, K. Sreenivasa*, M. Schenkel*, N. Chamachi*, P. Schake*, G. Krainer, M. Schlierf$
BioRxiv
Software on github
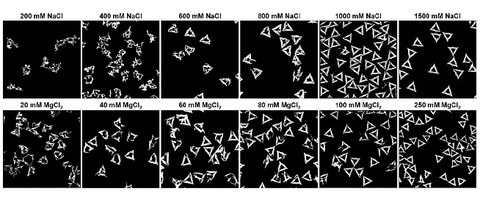
Recruiting Unicellular Algae for the Mass Production of Nanostructured Perovskites
L. Kuhrts, L. Helmbrecht, W. L. Noorduin, D. Pohl, X. Sun, A. Palatnik, C. Wetzker, A. Jantschke, M. Schlierf, I. Zlotnikov
Advanced Science
SI
2022
Single-molecule approaches reveal outer membrane protein biogenesis dynamics
A. Svirina, N. Chamachi, M. Schlierf
BioEssays
SI
Impact of cholesterol and Lumacaftor on the folding of CFTR helical hairpins
M. Schenkel*, D. Ravamehr-Lake*, T. Czerniak, J. P. Saenz, G. Krainer, M. Schlierf, C. M. Deber
BBA Biomembranes
Reliability and accuracy of single-molecule FRET studies for characterization of structural dynamics and distances in proteins
G. Agam, C. Gebhardt, M. Popara, R. Mächtel, J. Folz, B. Ambrose, N. Chamachi, S. Yoon Chung, et al.
bioRxiv
SI
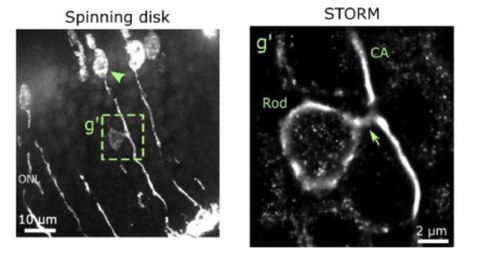
Evidence for endogenous exchange of cytoplasmic material between a subset of cone and rod photoreceptors within the adult mammalian retina via direct cell-cell connections
P. Heisterkamp*, O. Borsch*, N. Diaz Lezama, S. Gasparini, A. Fathima, L. S. Carvalho, F. Wagner, M. O. Karl, M. Schlierf, M. Ader
Exp Eye Research
SI
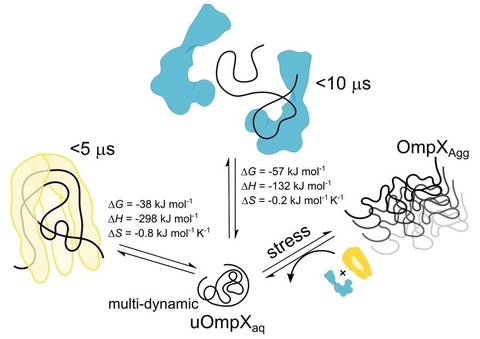
Chaperones Skp and SurA dynamically expand unfolded OmpX and synergistically disassemble oligomeric aggregates
N. Chamachi*, A. Hartmann*, M. Quynh Ma, A. Svirina, G. Krainer$, M. Schlierf$
PNAS
SI
2021
Heat treatment of thioredoxin fusions increases the purity of alpha-helical transmembrane protein constructs
M. Schenkel*, A. Treff, C. M. Deber, G. Krainer$, M. Schlierf$
Protein Science
SI
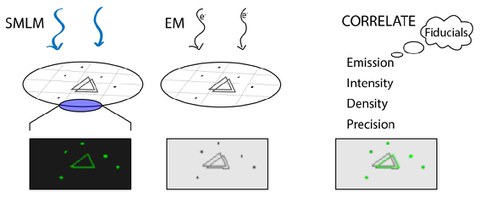
Precision of fiducial marker alignment for correlative super‐resolution fluorescence and transmission electron microscopy
A. Fathima*, C. Augusto Quintana-Cataño, C. Heintze, M. Schlierf$
Discover Materials
SI
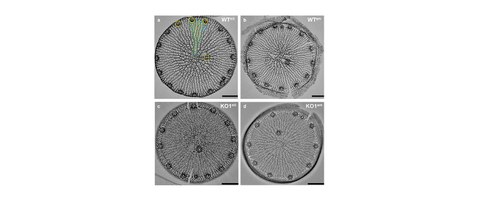
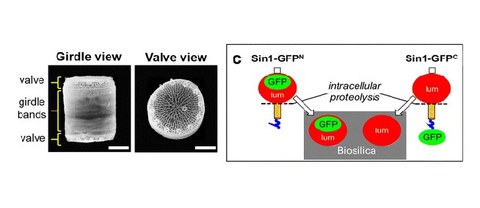
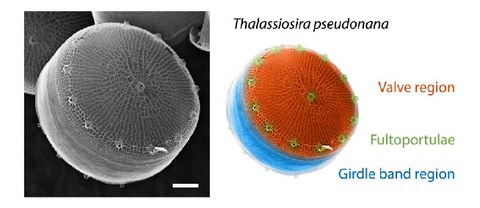
Computational analysis of the effects of nitrogen source and sin1 knockout on biosilica morphology in the model diatom Thalassiosira pseudonana
S. Horvát*, A. Fathima*, S. Görlich, M. Schlierf$, C. D. Modes$, N. Kröger$
Discover Materials
SI
2020
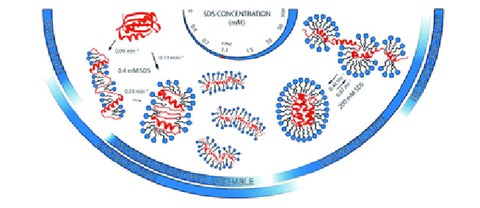
SDS-induced multi-stage unfolding of a small globular protein through different denatured states revealed by single-molecule fluorescence
G. Krainer*$, A. Hartmann*, V. Bogatyr*, J. Nielsen, M. Schlierf$, DE. Otzen$
Chemical Science
SI
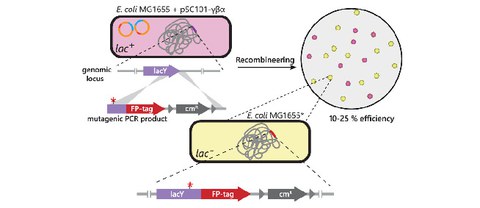
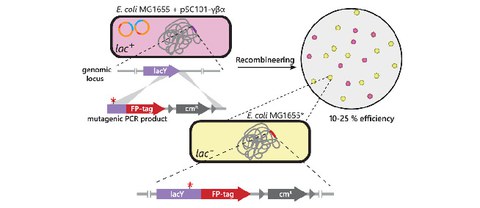
Fast, simultaneous tagging and mutagenesis of genes on bacterial chromosomes
L. Schärfen, M. Tišma, M. Schlierf
ACS Synthetic Biology
SI
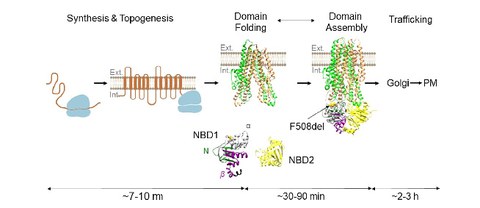
Towards next generation therapies for cystic fibrosis: Folding, function and pharmacology of CFTR
SJ. Bose, G. Krainer, DRS. Ng, M. Schenkel, H. Shishido, JS. Yoon, PM, Haggie, M. Schlierf, DN. Sheppard, WR Skach
Journal of Cystic Fibrosis
CFTR transmembrane segments are impaired in their conformational adaptability by a pathogenic loop mutation and dynamically stabilized by Lumacaftor
G. Krainer*$, M. Schenkel*, A. Hartmann, D. Ravamehr-Lake, CM. Deber$, M.Schlierf$
2019
Real-time monitoring of protein-induced DNA conformational changes using single-molecule FRET
L. Schärfen, M. Schlierf$
Methods 169, 11-20
Enhancing the stability of DNA origami nanostructure: staple strand redesign versus enzymatic ligation
S. Ramakrishnan*, L. Schärfen*, K. Hunold*, S. Fricke, G. Grundmeier, M. Schlierf$, A. Keller$, G. Krainer$
Nanoscale 11, 16270
SI
Structural dynamics of membrane-protein folding from single-molecule FRET
G. Krainer, S. Keller, M. Schlierf
Curr Opin Struc Biol (2019) 58: 124-137
SI
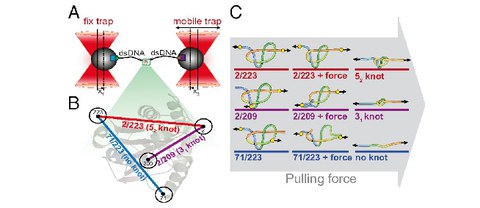
Structural heterogeneity of attC integron recombination sites revealed by optical tweezers
A. Mukhortava*, M. Pöge*, M.S. Grieb, A. Nivina, C. Loot, D. Mazel, M.Schlierf$
Nucleic Acids Research 47 (4), 1861–1870
SI
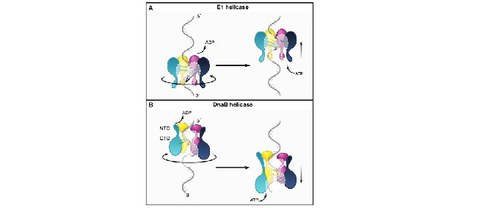
Hexameric helicase G40P unwinds DNA in single base pair steps
M. Schlierf$, G. Wang, X.S. Chen, T. Ha$
eLife 2019, 42001
2018
A Minimal Helical-Hairpin Motif Provides Molecular-Level Insights into Misfolding and Pharmacological Rescue of CFTR
G. Krainer*$, A. Treff*, A. Hartmann, T.A. Stone, M. Schenkel, C.M. Deber$, M. Schlierf$
Communications Biology 2018, 1, 154
SI
Conformational Dynamics Govern the Free-Energy Landscape of a Membrane-Interacting Protein
E. Frotscher*, G. Krainer*, A. Hartmann, M. Schlierf$, S. Keller$
ACS Omega 2018 3 (9), pp 12026–12032
SI
Precision and accuracy of single-molecule FRET measurements - a worldwide benchmark study
B. Hellenkamp, S. Schmid, O. Doroshenko, O. Opanasyuk, R. Kühnemuth, SR. Adariani, A. Barth, V. Birkedal, et al.
Nature Methods 15(9) 669-676 see also arXiv 1710.03807
SI
ATPase and Protease Domain Movements in the Bacterial AAA+ Protease FtsH Are Driven by Thermal Fluctuations
M. Ruer*, G. Krainer*, P. Gröger, M. Schlierf
JMB 2018, 430(22) 4592 see also biorxiv 323055
SI
Dissecting Nanosecond Dynamics in Membrane Proteins with Dipolar Relaxation upon Tryptophan Photoexcitation
E. Frotscher, G. Krainer, M. Schlierf, and S. Keller
Journal of Physical Chemistry Letters (2018) 9, 2241-2245
In situ temperature monitoring in single-molecule FRET experiments
A. Hartmann*, F. Berndt*, S. Ollmann*, G. Krainer, M. Schlierf
The Journal of Chemical Physics (2018) 148, 123330
SI
Ultrafast protein folding in membrane-mimetic environments
G. Krainer, A. Hartmann, A. Anandamurugan, P. Gracia, S. Keller, and M. Schlierf
Journal of Molecular Biology (2018) 430(4), 554-564
SI
2017
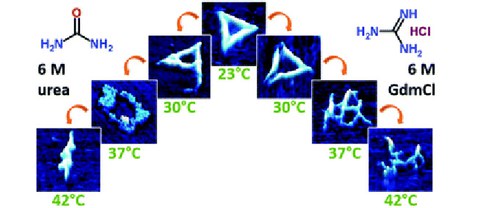
Cation-induced stabilisation and denaturation of DNA origami nanostructures in urea and guanidinium chloride
S. Ramakrishnan, G. Krainer, G. Grundmeier, M. Schlierf, A. Keller
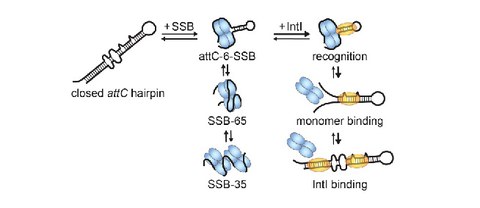
Dynamic stepwise opening of integron attC DNA hairpins by SSB prevents toxicity and ensures functionality
M.S. Grieb, A. Nivina, B.L. Cheeseman, A. Hartmann, D. Mazel, and M. Schlierf
Nucleic Acids Research 45 (18) 10555–10563 (open access)
SI
Silicanin-1 is a conserved diatom membrane protein involved in silica biomineralization
A. Kotzsch, P. Gröger, D. Pawolski, P. H. H. Bomans, N. A. J. M. Sommerdijk, M. Schlierf, N. Kröger
Forbidden chemistry: two-photon pathway in [2+2] cycloaddition of maleimides
M.V. Tsurkan, C. Jungnickel, M. Schlierf, and C. Werner
Journal of the American Chemical Society (2017), 139 (30), pp 10184–10187
SI
Slow interconversion in a heterogeneous unfolded-state ensemble of Outer Membrane Phospholipase A
G. Krainer *, P. Gracia *, E. Frotscher *, A. Hartmann, P. Gröger, S. Keller$ and M. Schlierf$
Biophysical Journal (2017), 113 (6), pp 1280-1289
SI
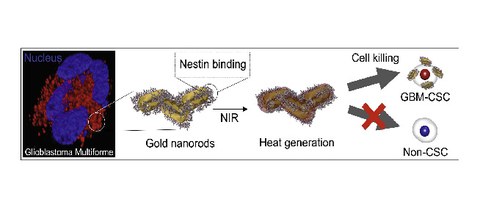
Enhanced targeting of invasive Glioblastoma cells by peptide-functionalized gold nanorods in hydrogel-based 3D cultures
D.P.N. Gonçalves, R.D. Rodriguez, T. Kurth, L.J. Bray, M. Binner, C. Jungnickel, F.N. Gür, S.W. Poser, T.L. Schmidt, D.R.T. Zahn, A. Androutsellis-Theotokis, M. Schlierf, C. Werner
Acta Biomaterialia (2017) 58, pp 12-25
SI
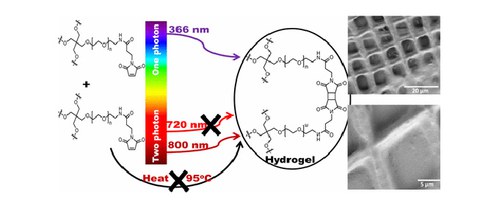
Bottom-Up Structuring and Site-Selective Modification of Hydrogels Using a Two-Photon [2+2] Cycloaddition of Maleimide
C. Jungnickel, M.V. Tsurkan, K. Wogan, C. Werner, M. Schlierf
Advanced Materials (2017) 29, 1603327
SI
2016
Establishing super-resolution imaging for proteins in diatom biosilica
P. Gröger, N. Poulsen, J. Klemm, N. Kröger, M. Schlierf
Scientific Reports (2016) 6, 36824
Knotting and unknotting of a protein in single molecule experiments
F. Ziegler, N.C.H. Lim, S.S. Mandal, B. Pelz, W.P. Ng, M. Schlierf, S.E. Jackson, M. Rief
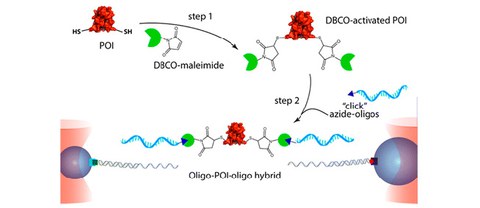
Efficient Formation of Site-specific Protein-DNA Hybrids using Copper-free Click Chemistry
A. Mukhortava$ and M. Schlierf$
Bioconjugate Chemistry 2016, 27 (7), pp 1559–1563
SI
Structural stability of DNA origami nanostructures in the presence of chaotropic agents
S. Ramakrishnan*, G. Krainer*, G. Grundmeier, M. Schlierf$, A. Keller$
Nanoscale 2016 8, 10398-10405
SI
Simultaneous single-molecule force and fluorescence sampling of DNA nanostructure conformations using magnetic tweezers
F.E. Kemmerich*, M. Swoboda*, D.J. Kauert, M.S. Grieb, S.Hahn, F.W. Schwarz, R. Seidel$, M. Schlierf$
Nano Letters, 2016, 16 (1), pp 381–386
SI
2015
Quantification of Millisecond Protein-Folding Dynamics in Membrane-Mimetic Environments by Single-Molecule FRET Spectroscopy
A. Hartmann*, G. Krainer*, S. Keller, M. Schlierf
Analytical Chemistry 2015, 87 (22), pp 11224–11232
SI
farFRET: Extending the Range in Single-Molecule FRET Experiments beyond 10 nm
G. Krainer*, A. Hartmann*, M. Schlierf
Nano Lett., 2015, 15 (9), pp. 5826-5829
SI
Diffusible Crosslinkers Generate Directed Forces in Microtubule Networks
Z. Lansky, M. Braun, A. Lüdecke, M. Schlierf, P. R. ten Wolde, M. E. Janson, S. Diez
Self-Assembling Hydrogels Crosslinked Solely by Receptor-Ligand Interactions: Tunability, Rationalization of Physical Properties and 3D Cell Culture
M. Thompson, M. Tsurkan, K. Chwalek, M. Bornhauser, M. Schlierf, C. Werner, Y. Zhang
Chem. Eur. J., 21(8), 3178-3182
SI
Bacterial initiators form dynamic filaments on single-stranded DNA monomer by monomer
H.-M. Cheng, P. Gröger, A. Hartmann, M. Schlierf
Nucleic Acid Research, 43(1), 396-405
SI
2014
Different Fluorophore Labeling Strategies and Designs Affect Millisecond Kinetics of DNA Hairpins
A. Hartmann, G. Krainer and M. Schlierf
Measuring Two at the Same Time: Combining Magnetic Tweezers with Single-Molecule FRET
M. Swoboda, M.S. Grieb, S. Hahn, M. Schlierf
Fluorescent Methods for Molecular Motors, Experientia Supplementum, 105, 253-276
2012
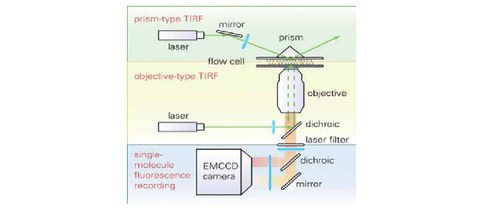
Studying molecular machines by TIRF microscopy
M. Schlierf and S. Diez
Biotech International Magazine, 5
A Helicase with an Extra Spring in Its Step
M. Schlierf and T. Ha
Enzymatic Oxygen Scavenging for Photostability without pH Drop in Single-Molecule Experiments
M. Swoboda, J. Henig, H.-M. Cheng, D. Brugger, D. Haltrich, N. Plumere and M. Schlierf
2010
Complex Unfolding Kinetics of Single-Domain Proteins in the Presence of Force
M. Schlierf, Z. T. Yew, M. Rief and E. Paci
Insight into helicase mechanism and function revealed through single-molecule approaches
J. G. Yodh, M. Schlierf and T. Ha
Q Rev Biophys., 43(2), 185-217
Direct evidence of the multidimensionality of the free-energy landscapes of proteins revealed by mechanical probes
Z. T. Yew, M. Schlierf, M. Rief and E. Paci
Drosophila Translational Elongation Factor-1g Is Modified in Response to DOA Kinase Activity and Is Essential for Cellular Viability
Y. Fan, M. Schlierf, et al.
Force–Fluorescence Spectroscopy at the Single-Molecule Level
R. Zhou, M. Schlierf and T. Ha
Methods in Enzymology, 475, 405-426
SI
2009
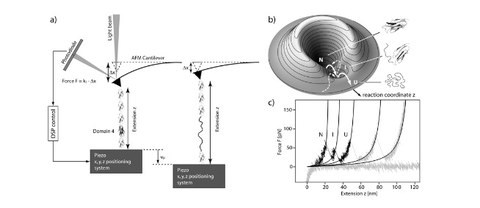
Surprising Simplicity in the Single-Molecule Folding Mechanics of Proteins
M. Schlierf and M. Rief
Angew. Chem. Int. Ed., 48(4), 820–822
2007
Direct Observation of Active Protein Folding Using Lock-in Force Spectroscopy
M. Schlierf, F. Berkemeier and M. Rief
2006
Mechanically controlled preparation of protein intermediates in single molecule experiments
F. Berkemeier, M. Schlierf and M. Rief
phys. stat. sol. (a) 203, No. 14, 3492–3495
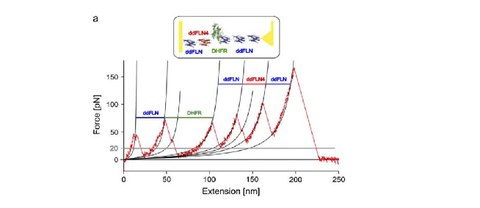
Fingerprinting DHFR in single-molecule AFM studies - Response to the comment by Ainavarapu et al.
M. Rief, J. P. Junker, M. Schlierf, K. Hell and W. Neupert
Cysteine engineering of polyproteins for singlemolecule force spectroscopy
H. Dietz, M. Bertz, M. Schlierf, F. Berkemeier, T. Bornschlögl, J. P. Junker and M. Rief
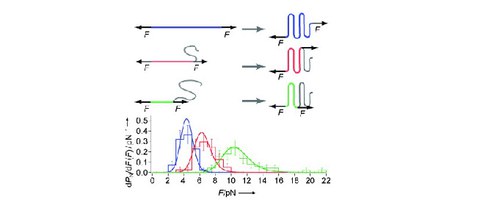
Single-Molecule Unfolding Force Distributions Reveal a Funnel-Shaped Energy Landscape
Michael Schlierf and Matthias Rief
2005
Temperature Softening of a Protein in Single-molecule Experiments
M. Schlierf and M. Rief
Influence of Substrate Binding on the Mechanical Stability of Mouse Dihydrofolate Reductase
J.P. Junker, K. Hell, M. Schlierf, W. Neupert and M. Rief
2004
The unfolding kinetics of ubiquitin captured with single-molecule force-clamp techniques
M. Schlierf, H. Li and J. M. Fernandez