Jul 23, 2025
Single-Cell Epigenetic Analysis Provides Insights Into Brain Development
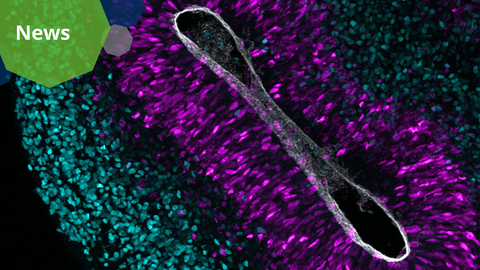
A microscopy picture of an 8-week-old human cortical organoid. Neurons are labeled in cyan and neural stem cells are labeled in magenta. The grey color shows the lining of one of the brain’s internal cavities.
Epigenetic regulation plays an important role in the development of most organs, but how important is it for the human brain? A research team led by Prof. Mareike Albert at the Center for Regenerative Therapies Dresden (CRTD) has now combined human brain organoids with a new single-cell technique to characterize a large number of epigenetic modifications in the developing brain. The team showed that histone methylation, a type of epigenetic modification, regulates cell proliferation and organoid size. The new method, called Epi-CyTOF, is now available for researchers and industry partners at the CRTD Mass Cytometry Facility.
Epigenetic modifications regulate gene expression in cells without altering the DNA sequence. These modifications, including post-translational modifications of histones, play an important role in organ development. However, their specific contribution to human brain development remains less understood.
“Mutations in epigenetic factors are known to cause different neurodevelopmental disorders. But what are the underlying molecular mechanisms? How do epigenetic factors regulate brain development?” says Mareike Albert, Professor at the Faculty of Biology and research group leader at the CRTD, TUD Dresden University of Technology. “One of the biggest challenges in answering these questions is the complex composition of the brain – it consists of many specialized cell types that emerge in different stages of development. On top of that, we have limited access to tissue samples of the human developing brain.”
To overcome this challenge, a team led by Prof. Albert combined two state-of-the-art tools: human brain organoids – lab-grown 3D structures that mimic the developing brain – and Epi-CyTOF, a powerful cytometry method that can analyze over 30 epigenetic modifications at single-cell resolution.
A Technological Leap
“Traditionally, researchers used Western blotting to detect global levels of various histone modifications,” explains Ezgi Şenoğlu, one of the authors of the study. “But this method has limitations. It is hard to scale, requires a lot of time and effort, and cannot distinguish cell types in complex tissues.”
Working with Dr. Claudia Peitzsch at the CRTD’s Mass Cytometry Facility, the Albert group instead applied Epi-CyTOF using a custom panel of more than 30 antibodies targeting different epigenetic modifications alongside 10 markers for different neural cell types. This allowed them to examine major neural cell types in the developing human brain and trace how epigenetic modifications change during neural stem cell differentiation.
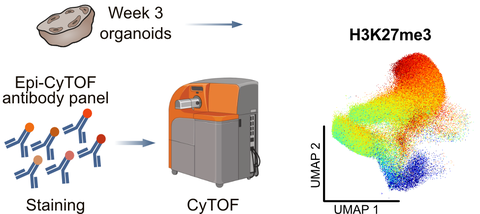
An overview of the experimental workflow and an example result of Epi-CyTOF.
Histone Methylation in the Focus
The team discovered that as the neural stem cells differentiate into neurons, the cells accumulate histone modifications, with histone 3 lysine 27 tri-methylation (H3K27me3) being the most strongly enriched mark. This modification was found on genes responsible for neocortex development and played a role in regulating neural stem cell proliferation and ultimately, in determining organoid size.
“Surprisingly, we also detected these modifications on genes that regulate the extracellular matrix – a complex network of proteins that surrounds the cells and helps create the stem cell niche,” says Nora Ditzer, one of the authors of the study. “These changes could indicate that epigenetic regulation shapes the environment of neural stem cells, an indirect role that could be crucial for brain development.”
It has been shown that mutations in genes controlling epigenetic regulation can disrupt normal brain development, leading to neurodevelopmental disorders that affect brain structure, function, and cognition.
“Mapping the timeline and targets of epigenetic modifications is crucial for understanding neurodevelopmental disorders and how gene expression is precisely controlled in development,” says Prof. Albert. “Since epigenetic regulation is very dynamic and reversible, the epigenome seems a promising target for therapeutic interventions.”
Funding
The research was funded by the BMFTR, as part of the ERA-NET NEURON MEPIcephaly consortium. A joint seed grant from the German Centers for Health Research (DZG) supported the setting up of the Epi-CyTOF panel for organoid research.
Innovative Technology Available for Everyone
The Epi-CyTOF method can be applied to other organs or disease models. The Mass Cytometry Facility at the CRTD offers support for experimental design, labeling, and data analysis to both researchers and industry partners. Anyone interested in the Epi-CyTOF or other mass cytometry technologies can contact the facility at .
Original Publication:
Nora Ditzer, Ezgi Senoglu, Theresa M. Schütze, Aikaterina Nikolaidi, Annika Kolodziejczyk,Katrin Sameith, Sevina Dietz, Razvan P. Derihaci, Cahit Birdir, Anne Eugster, Mike O. Karl, Andreas Dahl, Pauline Wimberge, Franziska Baenke, Claudia Peitzsch, Mareike Albert: Epigenome profiling identifies H3K27me3 regulation of extra-cellular matrix composition in human corticogenesis. Neuron (July 2025)
Link: https://doi.org/10.1016/j.neuron.2025.06.016
