Cannibalism in Bacillus subtilis colonies: Role of programmed cell death in shaping and functionalizing differentiated multicellular populations
- Prof Dr Klaus Dreisewerd Westfälische Wilhems-Universität Münster, Institut für Hygiene, Forschungsabteilung Biomedizinische Massenspektrometrie.
- Prof Dr Thorsten Mascher Technische Universität Dresden, Institut für Mikrobiologie, Arbeitsgruppe, Allgemeine Mikrobiologie.
Cannibalism is a unique bacterial form of programmed cell death (PCD) that links multicellular differentiation to endospore formation in Bacillus subtilis. On solid surfaces, undomesticated strains form intricately structured colonies, in which phenotypically different cell types are spatiotemporally coordinated to form robust microbial tissues and fruiting bodies. These structures provide a multicellular form for controlling population expansion, balancing growth with survival, and ultimately culminating in the formation of dormant endospores.
Cannibalism is based on the production of at least three toxins, the sporulation killing factor SKF, the sporulation delay protein SDP and the epipeptide EPE. Cannibalism seems to represent a central checkpoint in tissue formation, since it (i) provides resources to either delay or ultimately fuel sporulation, (ii) structures and functionalizes the colonies, and might ultimately serve to (iii) balance the ratio of different cell types in structured multicellular populations.
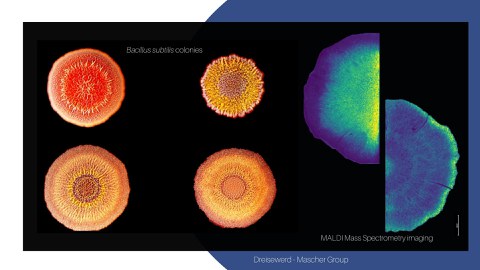
In this joint project, we aim to unravelling the role of PCD, mediated by cannibalism toxins and other secondary metabolites, for the structure and function of differentiated B. subtilis colonies. We aim at mechanistically understanding cannibalism toxin action both in 2D microcolonies at single-cell resolution and in 3D differentiated macrocolonies (Mascher). Cannibalism toxin production in colonies will also serve as a biological model system to apply and develop microbial MALDI mass spectrometry imaging (MSI), a core technology of this priority program. This powerful technique will be developed to enable studying the complex chemistry of bacterial colonies (cm range) in time and 3D space at single cell resolution (µm range).
Combining MSI with further imaging modalities, including life cell imaging, multi-parameter flow cytometry and single cell transcriptomic, will allow obtaining a comprehensive spatiotemporal map of microbial tissue dynamics and structure at unprecedented resolution. For developing MALDI-MSI (Dreisewerd), the current set-up for 2D top view and cross-section analysis will be optimized and the range of compounds detected will be expanded. A protocol for incorporating transmission geometries will be established by analyzing vertical thin-sections of colonies at µm resolution. Combined with 2D top view, this will allow analyzing bacterial colonies in 3D at high resolution down to single-cell level. Moreover, the established MALDI-MSI procedures shall be applied beyond B. subtilis, e.g. for studying PCD in E. coli and analyzing chemical communication in P. aeruginosa biofilms.
| Researchers |
| Jan-Philipp Knepper (PhD Student) |
| Lena Friebel (PhD Student) |
Contact Us
Westfälische Wilhems-Universität Münster
Email klaus.dreisewerd@uni-muenster.de
Tel +49 251 835 6726
Technische Universität Dresden
Email thorsten.mascher@tu-dresden.de
Tel +49 351 463 40420
Technische Universität Dresden
Email lena.friebel@tu-dresden.de
Tel +49 351 463 34511
Westfälische Wilhems-Universität Münster
Email jan-philipp.knepper@ukmuenster.de
Tel + 49 251 835 6274