Single-cell RNA sequencing of multicellular communities
- Prof Dr Susanne Häußler Twincore, a joint venture between Medical School Hannover (MHH) and Helmholtz Centre for Infection Research (HZI), Institute for Molecular Bacteriology.
Janne G Thöming
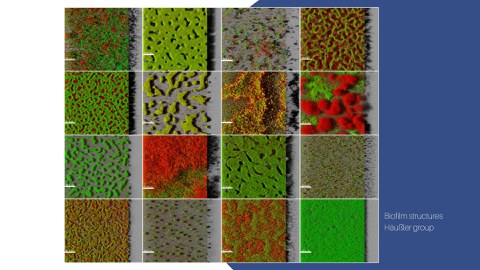
Multicellular bacterial communities exhibit various forms of heterogeneity, including co-occurrence of cells with different morphological traits, biochemical properties, and gene expression profiles.
Furthermore, in biofilm-associated infections, the genetic diversity of the infecting strain is a typical feature. This heterogeneity has been associated with higher productivity and lower susceptibility of the bacterial community to environmental changes, including antibiotic treatment as well as altered virulence traits. Due to the need and importance of a better understanding of this heterogeneity, methods using e.g. fluorescent reporters or probes to examine a specific gene of interest have been extensively applied.
More recently, single-cell RNA sequencing (scRNA-seq) has become a powerful tool for examining the heterogeneity of eukaryotic cells. This advanced technology is only at the beginning of its application for bacteria, as there are still some hurdles to overcome. The bacterial cell wall is incomparably more resistant to cell lysis, the mRNA has a shorter half-life and a much lower copy number, and isolating single cells from a multicellular community is a challenge. Nevertheless, the first recent work has provided valuable insight into factors influencing microbial heterogeneity.
In this proposal, we will establish single cell sequencing based on published protocols and study how the microenvironment and genetic variability of Pseudomonas aeruginosa biofilm population shape subpopulations and drive antibiotic tolerant phenotypes. We furthermore aim to contribute to the establishment of an SPP technology pipeline that involves single cell transcriptomics and FACS-based cell sorting. The integration of the data promises a detailed characterization of bacterial subpopulations of three model species (P. aeruginosa, Bacillus subtilis and Escherichia coli).
About Us
We are a multidisciplinary research group with the overall aim of uncovering genetic determinants of Pseudomonas aeruginosa pathogenicity, adaptation to a chronic, persistent state of infection, and recalcitrance towards antimicrobial treatment.
For this purpose, we apply high-throughput -omics techniques on a plethora of clinical P. aeruginosa isolates and use the generated large-scale sequence variation and gene expression datasets for genome-wide association studies.
With this approach, we aim to advance molecular diagnostics for resistance profiling and to develop alternative therapeutic strategies to combat chronic biofilm-associated infections.
| Researchers |
|
Dr Janne Gesine Thöming (Postdoc) Dr Jelena Erdmann (Postdoc) |
Visit Us
The Molecular Bacteriology Group is based at the Helmholtz Centre for Infection Research (HZI) in Braunschweig, at the TWINCORE in Hannover, and at the University Hospital (Rigshospitalet) in Copenhagen, Denmark.
Contact Us
Helmholtz Centre for Infection Research (HZI)
Email: susanne.haeussler@helmholtz-hzi.de
Tel: +49 531 6181 3000
Helmholtz Centre for Infection Research (HZI)
Email: jelena.erdmann@helmholtz-hzi.de
Tel: +49 531 6181 3000
Helmholtz Centre for Infection Research (HZI)
Email: janne.thoeming@helmholtz-hzi.de
Tel: +49 531 6181 3000
