Publications
LIST OF PEER-REVIEWED RESEARCH ARTICLES:
Junker, F.; Rupf, S.; Schindler, P.; Wilden, C.; Hohl, M.; Ruiz-Gómez, G.; Pisabarro, M.T.; Wrublewsky, S.; Bickelmann, C.; Berhorst, C.; Alansary, D.; Wieland, B.; Bischoff, M.; Lee, P. S.; Moeller, S.; Berg, A.; Dancker, T.; Lauterbach, M.A.; Ganse, B.; Prates Roma, L.; Steudter, T.; Metzger, W.; Tschernig, T.; Ampofo, E.; Laschke, M. W.; Hannig, M.; Rother, S. Glycosaminoglycan-functionalized hydrogels for sustained delivery of tissue inhibitor of metalloproteinase-3 mediating matrix metalloprotease inhibition and extracellular matrix stabilization.
Bioactive Materials 61:172-193 (2026)
https://authors.elsevier.com/sd/article/S2452199X2600071X
Kuhne, K.; Strohbach, L.; Neuber, C.; Wodtke, R.; Ruiz-Gómez, G.; Belter, B.; Brandt, F.; Gluhacevic von Krüchten, L.; Keller, M.; Pisabarro, M. T.; Kopka, K.; Pietzsch, J.; Löser, R. Development of cathepsin B-activatable cell-penetrating peptides for tumor targeting.
ACS Pharmacology & Translational Science accepted (2026)
Ruiz-Gómez, G.; Uvizl, A.; Bakos, G.; Leung, J.; Pisabarro, M. T.; Mansfeld, J. De novo designed APC/C inhibitors provide a rationale for targeting RING-type E3 ubiquitin ligases. Journal of Medicinal Chemistry 68(11):11468-11483 (2025)
https://doi.org/10.1021/acs.jmedchem.5c00416
Wrublewsky, S.; Rother, S.; Pohlemann, F.; Radanovic, T.; Junker, F.; Boewe, A. S.; Schunk, S.; Roma, L. P.; Ruiz-Gómez, G.; Pisabarro, M. T.; MacDonald, P. E.; Menger, Michael, D.; Laschke, M. W.; Ampofo, E. Heparan sulfate fine-tuned interleukin (IL)-1 signaling inhibits insulin secretion of grafted pancreatic islet.
Science Advances 11(32):eady8566 (2025)
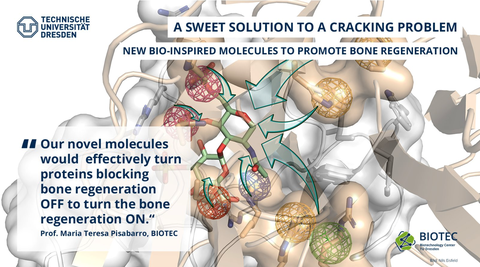
Kramp, P.; Özmaldar, A.; Ruiz-Gómez, G.; Pisabarro, M. T. In silico analysis of serum albumin binding by bone-regenerative hyaluronan-based molecules.
Pharmaceutics 17:1445 (2025)
Hoersten J.; Ruiz-Gómez, G.; Paszkowski-Rogacz, M.; Gilioli, G.; Guillem-Gloria, P. M.; Lansing, F.; Pisabarro, M. T.; Buchholz, F. Engineering spacer specificity of the Cre/loxP system.
Nucleic Acids Research 52(13):8017-8031 (2024)
Döring, M.; Brux, M.; Paszkowski-Rogacz, M.; Guillem-Gloria, P. M.; Buchholz, F.; Pisabarro, M. T.; Theis, M. Nucleolar protein TAAP1/C22orf46 confers pro-survival signaling in non-small cell lung cancer.
Life Science Alliance 7(4):e202302257 (2024)
Ruiz-Gómez, G.; Salbach-Hirsch J.; Dürig, J. N.; Köhler, L.; Balamurugan, K.; Rother, S.; Heidig, S. L.; Moeller, S.; Schnabelrauch, M.; Furesi, G.; Pählig, S.; Guillem-Gloria, P. M.; Hofbauer, C.; Hintze, V.; Pisabarro, M. T.; Rademann, J.; Hofbauer, L. C. Rational engineering of glycosaminoglycan-based Dickkopf-1 scavengers to improve bone regeneration.
Biomaterials 297:122105 (2023)
A Sweet Solution to a Cracking Problem: Scientists Design New Bio-Inspired Molecules to Promote Bone Regeneration.
Biomaterials 297:122105 (2023)
Behring, L.;A.; Ruiz-Gómez, G.; Trapp, C.; Morales, M.; Wodtke, R.; Köckerling, M.; Kopka, K.; Pisabarro, M. T.; Pietzsch, J.; Löser, R. Dipeptide-derived alkynes as potent and selective irreversible inhibitors of cysteine cathepsins.
Journal of Medicinal Chemistry 66(6):3818-3851 (2023)
Guillén-Pingarrón, C.; Guillem-Gloria, P. M.; Soni, A.; Ruiz-Gómez, G.; Ausburg, M.; Buchholz, F.; Anselmi, M.; Pisabarro, M. T. Conformational dynamics promotes disordered regions from function-dispensable to essential in evolved site-specific DNA recombinases. Computational and Structural Biotechnology Journal 20:989-1001 (2022)
Hoersten, J.; Ruiz-Gómez, G.; Lansing, F.; Rojo-Romanos, T.; Schmitt, L. T.; Sonntag, J.; Pisabarro, M. T.; Buchholz, F. Pairing of single mutations yields obligate Cre-type site-specific recombinases.
Nucleic Acids Research 50(2):1174-1186 (2022)
Müller, C. D.; Ruiz-Gómez, G.; Cazzonelli, S.; Möller, S.; Wodtke, R.; Löser, R.; Freyse, J.; Dürig, J-N.; Rademan, J.; Hempel, U.; Pisabarro, M. T.; Vogel, S. Sulfated glycosaminoglycans inhibit transglutaminase 2 by stabilizing its closed conformation.
Scientific Reports 12:13326 (2022)
Balamurugan, K.; Koehler, L.; Dürig, J.N.; Hempel, U.; Rademann, J.; Hintze, V.; Pisabarro, M. T. Structural insights into the modulation of PDGF/PDGFR-β complexation by hyaluronan derivatives.
Biological Chemistry 402(11):1441-1452 (2021)
Balamurugan, K. and Pisabarro, M. T. Stabilizing Role of Water Solvation on Anion-pi Interactions in Proteins.
ACS Omega 6, 39:25350-25360 (2021)
Rother, S.; Ruiz-Gómez, G.; Balamurugan, K.; Koehler, L.; Fiebig, K. M.; Galiazzo, V. D.; Hempel, U.; Moeller, S.; Schnabelrauch, M.; Waltenberger, J.; Pisabarro, M. T.; Hintze, V. Scharnweber, D. Hyaluronan/collagen hydrogels with sulphated glycosaminoglycans maintain VEGF165 activity and fine-tune endothelial cell response.
ACS Applied Bio Materials 4(1):494-506 (2021)
Soni, A.; Ausburg, M.; Buchholz, F.; Pisabarro, M. T. Nearest-neighbor amino acids of specificity-determining residues influence the activity of engineered Cre-type recombinases.
Scientific Reports, 10:13985 (2020)
Koehler, L.; Ruiz-Gómez, G.; Balamurugan, K.; Rother, S.; Freyse, J.; Möller, S.; Schnabelrauch, M.; Köhling, S.; Djordjevic, S.; Scharnweber, D.; Rademann, J.; Pisabarro, M. T.; Hintze, V. Dual action of sulfated hyaluronan on angiogenic processes in relation to vascular endothelial growth factor-A.
Scientific Reports, 9(1):18143 (2019)
Ruiz-Gómez, G.; Vogel, S.; Möller, S.; Pisabarro, M. T.; Hempel, U. Glycosaminoglycans influence enzyme activity of MMP2 and MMP2/TIMP3 complex formation- Insights at celular and molecular level.
Scientific Reports, 9:4905 (2019)
Köling, S.; Blaszkiewicz, J.; Ruiz-Gomez, G.; Fernández-Bachiller, M. I.; Lemmnitzer, K.; Panitz, N.; Beck-Sickinger, A. G.; Schiller, J.; Pisabarro, M. T.; Rademann, J. Syntheses of defined sulfated oligohyaluronans reveal structural effects, diversity and thermodynamics of GAG-protein binding.
Chemical Science, 10:866-878 (2019)
Thönes, S.; Rother, S.; Wippold, T.; Blaszkiewicz, J.; Balamurugan, K.; Moeller, S.; Ruiz-Gómez, G.; Schnabelrauch, M.; Scharnweber, D.; Saalbach, A.; Rademann, J.; Pisabarro, M. T.; Hintze, V.; Anderegg, U. Hyaluronan/collagen hydrogels containing sulfated hyaluronan improve wound healing by sustained release of Heparin-Binding EGF-like growth factor.
Acta Biomaterialia, 86:135-147 (2019)
Wodtke, R.; Hauser, C.; Ruiz-Gomez, G.; Jäckel, E.; Bauer, D.; Lohse, M.; Wong, A.; Pufe, J.; Ludwig, F.; Fischer, S.; Hauser, S.; Greif, D.; Pisabarro, M. T.; Pietzsch, J.; Pietsch, M.; Löser, R. Ne -Acryloyllysine piperazides as irreversible inhibitors of transglutaminase 2 – synthesis, structure-activity relationships and pharmacokinetic profiling.
Journal of Medicinal Chemistry, 61(10):4528-4560 (2018)
Koehler, L.; Samsonov, S.; Rother, S.; Vogel, S.; Köling, S.; Moeller, S.; Schnabelrauch, M.; Rademann, J.; Hempel, U.; Pisabarro, M. T.; Scharnweber, D.; Hintze, V. Sulfated hyaluronan derivatives modulate TGF-β1:receptor complex formation: Possible consequences for TGF-β1 signalling.
Scientific Reports, 7(1):1210 (2017)
Jouy, F.; Lohmann, N.; Wandel, E.; Ruiz-Gómez, G.; Pisabarro, M. T.; Beck-Sickinger, A. G.; Schnabelrauch, M.; Moeller, S.; Simon, J. C.; Kalkhof, S.; von Bergen, M.; Franz, S. Sulfated hyaluronan attenuates inflammatory signaling pathways in macrophages involving induction of antioxidants.
Proteomics, 17, 10:1700082 (2017)
Rother, S.; Samsonov, S.; Möller, S.; Schnabelrauch, M.; Rademann, J.; Blaszkiewicz, J.; Köhling, S.; Waltenberger, J.; Pisabarro, M. T.; Scharnweber, D.; Hintze, V. Sulfate hyaluronan alters endothelial cell activation in vitro by controlling the biological activity of the angiogenic factors vascular endothelial growth factor-A and tissue inhibitor of metalloproteinase-3.
ACS Appl Mater Interfaces, 9(11):9539-9550 (2017)
Babik, S.; Samsonov, S.A.; Pisabarro, M. T. Computational drill down on FGF1-heparin interactions through methodological evaluation.
Glycoconjugate Journal, 34(3): 427-440 (2017)
Ruiz-Gómez, G.; Hawkins, J. C.; Philipp, J.; Künze, G.; Wodtke, R.; Löser, R.; Fahmy, K.; Pisabarro, M. T. Rational structure-based rescaffolding approach to de novo design of interleukin 10 (IL-10) receptor-1 mimetics.
Plos One,11(4):e0154046 (2016)
Meinke, G.; Bohm, A.; Hauber, J.; Pisabarro, M. T.; Buchholz, F. Cre recombinase and other tyrosine recombinases.
Chemical Reviews,116(20):12785-12820 (2016)
Karpinski, J.; Hauber, I.; Chemnitz, J.; Schäfer, C.; Paszkowski-Rogacz, M.; Chakraborty, D.; Beschorner, N.; Hofmann-Sieber, H.; Lange, U. C.; Grundhoff, A.; Hackmann, K.; Schrock, E.; Abi-Ghanem, J.; Pisabarro, M. T.; Surendranath, V.; Schambach, A.; Lindner, C.; van Lunzen, J.; Hauber, J.; Buchholz, F. Directed evolution of a recombinase that excises the provirus of most HIV-1 primary isolates with high specificity.
Nature Biotechnology, 34(4):401-409 (2016)
Karimova, M.; Splith, V.; Karpinski, J.; Pisabarro, M. T.; Buchholz, F. Discovery of Nigri/nox and Panto/pox site-specific recombinase systems facilitates advanced genome engineering.
Scientific Reports, 6:30130 (2016)
Samsonov, S. A. and Pisabarro, M. T. Computational analysis of interactions in structurally available protein-glycosaminoglycan complexes.
Glycobiology, 26(8):850-861 (2016)
Rother, S.; Samsonov, S. A.; Hofmann, T; Blaszkiewicz, J.; Köhling, S.; Moeller, S.; Schnabelrauch, M.; Rademann, J.; Kalkhof, S; von Bergen, M; Pisabarro, M. T.; Scharnweber, D; Hintze, V. Structural and functional insights into the interaction of sulfated glycosamino-glycans with tissue inhibitor of metalloproteinase-3 - a possible regulatory role on extracellular matrix homeostasis.
Acta Biomaterialia, 45:143-154 (2016)
Rother, S.; Samsonov, S. A.; Hempel, U.; Vogel, S.; Möller, S.; Blaszkiewicz, J.; Köhling, S.; Schnabelrauch, M.; Rademann, J.; Pisabarro, M. T.; Hintze, V.; Scharnweber, D. Sulfated hyaluronan alters the interaction profile of TIMP-3 with the endocytic receptor LRP-1 cluster II and IV and increases the extracellular TIMP-3 level of human bone marrow stromal cells.
Biomacromolecules, 17(10):3252-3261 (2016)
Picke, A.-K.; Salbach-Hirsch, J.; Hintze, V.; Rother, S.; Rauner, M.; Kascholke, C.; Moeller, S.; Bernhardt, R.; Rammelt, S.; Pisabarro, M. T.; Ruiz-Gómez, G.; Schnabelrauch, M.; Schulz-Siegmund, M.; Hacker, M. C.; Scharnweber, D.; Hofbauer, C.; Hofbauer, L. C. Sulfated hyaluronan improves bone regeneration of diabetic rats by prolonged binding of sclerostin and enhancing osteoblast function.
Biomaterials, 96:11-23 (2016)
Panitz, N.; Theisgen, S.; Samsonov, S. A.; Gehrcke, J. P.; Baumann, L.; Bellmann-Sickert, K.; Pisabarro, M. T.; Rademann, J.; Huster, D.; Beck-Sickinger, A. G. The structural investigation of glycosaminoglycan binding to CXCL12 displays distinct interaction sites.
Glycobiology, 26(11):1209-1221 (2016)
Gehrcke, J. P. and Pisabarro, M. T. Identification and characterization of a glycosaminoglycan binding site on Interleukin-10 via molecular simulation methods.
Journal of Molecular Graphics and Modelling, 62:97-104 (2015)
Abi-Ghanem, J.; Samsonov, S. A.; Pisabarro, M. T. Insights into the preferential order of strand exchange in the Cre/loxP recombinase system: impact of the DNA spacer flanking sequence and flexibility.
Journal of Computer-Aided Molecular Design, 29(3):271-282 (2015)
Salbach-Hirsch, J.; Samsonov, S. A.; Hintze, V.; Hofbauer, C.; Picke, A.-K.; Rauner, M.; Gehrcke, J.-P.; Moeller, S.; Schnabelrauch, M.; Scharnweber, D.; Pisabarro, M. T.; Hofbauer, L. C. Structural and functional insights into sclerostin-glycosaminoglycan interactions in bone.
Biomaterials, 67:335-345 (2015)
Anselmi, M. and Pisabarro, M. T.Exploring multiple binding modes using confined replica exchange molecular dynamics.
Journal of Chemical Theory and Computation, 11(8):3906-3918 (2015)
Scharnweber, D.; Hübner, J.; Rother, S.; Hempel, U.; Anderegg, U.; Samsonov, S. A.; Pisabarro, M. T.; Hofbauer, L.; Schnabelrauch, M.; Franz, S.; Simon, S.; Hintze, V. Glycosaminoglycan derivatives - promising candidates for the design of functional materials.
Journal of Materials Science: Materials in Medicine, 26(9):232 (2015)
Hofmann, T.; Samsonov, S. A.; Pichert, A.; Lemmnitzer, K.; Schiller, J.; Huster, D.; Pisabarro, M. T.; von Bergen, M.; Kalkhof, S. Structural analysis of Interleukin-8 / Glycosaminoglycan interactions by amide hydrogen/deuterium exchange mass spectrometry.
Methods, 89:45-53 (2015)
Andrault, P. M.; Samsonov, S. A.; Weber, G.; Coquet, L.; Nazmi, K.; Bolscher, J.; Lalmanach , A. C.; Jouenne, T.; Bromme, D.; Pisabarro, M. T.; Lalmanach, G.; Lecaille, F. Antimicrobial peptide LL-37 is both a substrate of cathepsins S and K and a selective inhibitor of cathepsin L.
Biochemistry, 54(17):2785-2798 (2015)
Wodtke, R.; Ruiz-Gómez, G.; Kuchar, M.; Pisabarro, M. T.; Novotná, P.; Urbanová, M.; Steinbach, J.; Pietzsch, J.; Löser, R. Cyclopeptides containing the DEKS motif as conformationally restricted collagen telopeptide analogues: synthesis and conformational analysis.
Organic & Biomolecular Chemistry, 13(6):1878-1896 (2015)
Samsonov, S. A.; Bichmann, L.; Pisabarro, M. T. Coarse-grained model of glycosaminoglycans.
Journal of Chemical Information and Modeling, 55(1):114-124 (2015)
Hintze, V.; Samsonov, S. A.; Anselmi, M.; Möller, S.; Becher, J.; Schnabelrauch, M.; Scharnweber, D.; Pisabarro, M. T. Sulfated glycosaminoglycans exploit the conformational plasticity of bone morphogenetic protein-2 (BMP-2) and alter the interaction profile with its receptor.
Biomacromolecules, 15(8):3083-3092 (2014)
Künze, G.; Gehrcke, J. P.; Pisabarro, M. T.; Huster, D. NMR characterization of the binding properties and conformation of glycosaminoglycans interacting with interleukin-10.
Glycobiology, 24(11):1036-1049 (2014)
Samsonov, S. A.; Gehrcke, J. P.; Pisabarro, M. T. Flexibility and explicit solvent in molecular dynamics-based docking of protein-glycosaminoglycan systems.
Journal of Chemical Information and Modelling, 54(2):582-592 (2014)
Atkovska, K.; Samsonov, S. A.; Paszkowski-Rogacz, M.; Pisabarro, M. T. Multipose binding in molecular docking.
International Journal of Molecular Sciences, 15(2):2622-2645 (2014)
Samsonov, S. A.; Theisgen, S.; Riemer, T.; Huster, D.; Pisabarro, M. T. Glycosaminoglycan monosaccharide blocks analysis by quantum mechanics, molecular dynamics and nuclear magnetic resonance.
BioMed Research International, 2014:808071 (2014)
Samsonov, S. A. and Pisabarro, M. T. Importance of IdoA and IdoA(2S) ring conformations in computational studies of glycosaminoglycan-protein interactions.
Carbohydrate Research, 381:133-137 (2013)
Abi-Ghanem, J.; Chusainow, J.; Karimova, M.; Spiegel, C.; Hofmann-Sieber, H.; Hauber, J.; Buchholz, F.; Pisabarro, M. T. Engineering of a target site-specific recombinase by a combined evolution- and structure-guided approach.
Nucleic Acids Research, 41(4):2394-2403 (2013)
Karimova, M.; Abi-Ghanem J.; Berger, N.; Surendranath, V.; Pisabarro, M. T.; Buchholz, F. Vika/vox, a novel efficient and specific Cre/loxP-like site-specific recombination system.
Nucleic Acids Research, 41(2):e37 (2013)
Salbach-Hirsch, J.; Kraemer, J.; Rauner, M.; Samsonov, S. A.; Pisabarro, M. T.; Moeller, S.; Schnabelrauch, M.; Scharnweber, D.; Hofbauer, L. C.; Hintze, V. The promotion of osteoclastogenesis by sulfated hyaluronan through interference with osteoprotegerin and receptor activator of NF-κB ligand/osteoprotegerin complex formation.
Biomaterials, 34(31):7653-7661 (2013)
van der Smissen, A.; Samsonov, S. A.; Hintze, V.; Scharnweber, D.; Moeller, S.; Schnabelrauch, M.; Pisabarro, M. T.; Anderegg, U. Artificial extracellular matrix composed of collagen I and high-sulfated hyaluronan interferes with TGFβ1 signaling and prevents TGFβ1 induced myofibroblast differentiation.
Acta Biomaterialia, 9(8):7775-7786 (2013)
Möbius, K.; Nordsieck, K.; Pichert, A.; Samsonov, S. A.; Thomas, L.; Schiller, J.; Kalkhof, S.; Pisabarro, M. T.; Beck-Sickinger, A. G.; Huster, D. Investigation of lysine side chain interactions of Interleukin-8 with Heparin and other glycosaminoglycans studied by a methylation-NMR approach.
Glycobiology, 23(11):1260-1269 (2013)
Sage, J.; Mallevre, F.; Barbarin-Costes, F.; Samsonov, S.; Gehrcke, J.-P.; Pisabarro, M. T.; Perrier, E.; Schnebert, S.; Roget, A.; Livache, T.; Nizard, C.; Lalmanach, G.; Lecaille, F. Binding of chondroitin 4-sulfate to cathepsin S regulates its enzymatic activity.
Biochemistry, 52(37):6487-6498 (2013)
Nordsieck, K.; Pichert, A.; Samsonov, S.A.; Thomas, L.; Berger, C.; Pisabarro, M. T.; Huster, D.; Beck-Sickinger, A.G. Residue 75 of Interleukin-8 is crucial for its Interactions with Glycosaminoglycans.
ChemBioChem, 13(17):2558-2566 (2012)
Tomczak, A.; Sontheimer, J.; Drechsel, D.; Hausdorf, R.; Gentzel, M.; Shevchenko, A.; Eichler, S.; Fahmy, K.; Buchholz, F.; Pisabarro, M. T. 3D profile-based approach to proteome-wide discovery of novel human chemokines.
PLoS One, 7(5):e36151 (2012)
Hawkins, J. C.; Zhu, H.; Teyra, J. ; Pisabarro, M. T. Reduced false positives in PDZ binding prediction using sequence and structural descriptors.
IEEE/ACM Transactions on Computational Biology and Bioinformatics, 9(5):1492-1503 (2012)
Pichert, A.; Samsonov, S.A.; Theisgen, S.; Thomas, L.; Baumann, L.; Schiller, J.; Beck-Sickinger, A.G.; Huster, D.; Pisabarro, M.T. Characterization of the Interaction of Interleukin-8 with Hyaluronan, Chondroitin Sulfate, Dermatan Sulfate, and Their Sulfated Derivatives by Spectroscopy and Molecular Modelling.
Glycobiology, 22(1):134-145 (2012)
Teyra, J.; Samsonov, S.A.; Schreiber, S.; Pisabarro, M. T. SCOWLP update: 3D classification of protein-protein, -peptide, -saccharide and -nucleic acid interactions, and structure-based binding inferences across folds.
BMC Bioinformatics, 12:398 (2011)
Samsonov, S.A.; Teyra, J.; Pisabarro, M. T. Docking glycosaminoglycans to proteins: analysis of solvent inclusion.
Journal of Computer-Aided Molecular Design, 25(5):477-489 (2011)
Tomczak, A. and Pisabarro, M. T. Identification of CCR2-binding features in Cytl1 by a CCL2-like chemokine model.
PROTEINS: Structure, Function and Bioinformatics, 79(4):1277-1292 (2011)
Teyra, J.; Hawkins, J.; Zhu, H.; Pisabarro, M. T. Studies on the inference of protein binding regions across fold space based on structural similarities.
PROTEINS: Structure, Function and Bioinformatics, 79(2):499-508 (2011)
Zhu, H. and Pisabarro, M. T. MSPocket: An Orientation Independent Algorithm for the Detection of Ligand Binding Pockets.
Bioinformatics, 27(3):351-358 (2011)
Slabicki, M.; Theis, M.; Krastev, D.; Samsonov, S.; Mundwiller, E.; Junqueira, M.; Paszkowski-Rogacz, M.; Teyra, J.; Heninger, A-K.; Poser, I.; Prieur, F.; Truchetto, J.; Confavreux, C.; Marelli, C.; Durr, A.; Camdessanche, J. P.; Brice, A.; Shevchenko, F.; Pisabarro, M. T.; Stevanin, G.; Buchholz, F. A genome-scale DNA repair RNAi screen identifies SPG48 as a novel gene associated with hereditary spastic paraplegia.
PLoS Biology, 8(6):e1000408 (2010)
Paszkowski-Rogacz, M.; Slabicki, M.; Pisabarro, M. T.; Buchholz, F. PhenoFam - gene set enrichment analysis through protein structural information.
BMC Bioinformatics, 11:254 (2010)
Palencia A.; Camara-Artigas A.; Pisabarro M.T.; Martinez J.C.; Luque I. Role of interfacial water molecules in proline-rich ligand recognition by the Src homology 3 domain of Abl.
J. Biol. Chem., 285(4):2823-2833 (2010)
Samsonov, S.A.; Salwiczek, M.; Anders, G.; Koksch, B.; Pisabarro, M. T. Fluorine in protein environments: A QM and MD study.
J. Phys. Chem. B., 113(51)16400-16408 (2009)
Samsonov, S. A.; Teyra, J.; Anders, G.; Pisabarro, M. T. Analysis of the impact of solvent on contacts prediction in proteins.
BMC Struct. Biol., 9(1):22 (2009)
Salwiczek, M.; Samsonov, S.; Vagt, T.; Nyakatura, E.; Fleige, E.; Numata, J.; Cölfen, H.; Pisabarro, M. T.; Koksch, B. Position Dependent Effects of Fluorinated Amino Acids on Hydrophobic Core Formation of a Heterodimeric Coiled Coil.
Chem. Eur. J., 15(31):7628-7636 (2009)
Ding, L.; Paszkowski-Rogacz, M.; Nitzsche, A.; Slabicki, M.; Heninger, A-K; Kittler, R. Junqueira, M.; Shevchenko, A.; Schulz, H.; Hubner, N.; Doss, M. X.; Sachinidis, A.; Hescheler, J.; Iacone, R.; Anastassiadis, K.; Stewart, F.; Pisabarro, M. T.; Caldarelli, A.; Poser, I.; Theis, M.; Buchholz, F. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity.
Cell Stem Cell, 4(5):403-515 (2009)
Theis, M.; Slabicki, M.; Junqueira, M.; Paszkowski-Rogacz, M.; Sontheimer, J.; Kittler, R.; Heninger, A.; Kruusmaa, K.; Poser, I.; Hyman, A.; Pisabarro, M. T.; Gstaiger, M.; Aebersold, R.; Shevchenko, A.; Buchholz, F. Comparative Profiling Identifies C13orf3 as a Novel Component of the Ska Complex Required for Mammalian Cell Division.
EMBO J., 28(10):1453-1465 (2009)
Vagt, T.; Jäckel, C.; Samsonov, S.; Pisabarro, M. T.; Koksch, B. Selection of a buried salt bridge by phage display.
Bioorg. Med. Chem. Lett., 19(14):3924-3927 (2009)
Samsonov, S.; Teyra, J.; Pisabarro, M. T. A molecular dynamics approach to study the importance of solvent in protein interactions.
PROTEINS: Structure, Function and Bioinformatics, 73(2):515-525 (2008)
Baldauf, C.; Pisabarro, M. T. Stable hairpins with beta-peptides: route to tackle protein-protein interactions.
J. Phys. Chem. B,112(25):7581-7591(2008)
Teyra, J.; Paszkowski-Rogacz, M.; Anders, G.; Pisabarro, M. T. SCOWLP classification: Structural comparison and analysis of protein binding regions.
BMC Bioinformatics, 9:9 (2008)
Lättig, J.; Böhl, M.; Fischer, P.; Tischer, S.; Tietböhl, C.; Menschikowski, M.; Gutzeit, H. O.; Metz, P.; Pisabarro, M. T. Mechanism of Inhibition of Human Secretory Phospholipase A2 by Flavonoids. Rationale for Lead Design.
Journal of Computer-Aided Molecular Design, 21(8):473-483 (2007)
Scheike, J.; Baldauf, C.; Spengler, J.; Albericio, F.; Pisabarro, M. T.; Koksch, B. Amide-to-ester substitution in coiled coils: the effect of H-bond elimination on protein structure formation.
Angewandte Chemie, 46(41):7766-7769 (2007)
Teyra, J. and Pisabarro, M. T. Characterization of Interfacial Solvent in Protein Complexes and Contribution of Wet Spots to the Interface Description.
PROTEINS: Structure, Function and Bioinformatics, 67, 1087-1095 (2007)
Pisabarro, M. T.; Leung, B; Kwong, M.; Corpuz, R.; Frantz, G. D.; Chiang, N.; Vandlen, R.; Diehl, L. J.; Skelton, N.; Eaton, D.; Schmidt, K. N. Novel Human Dendritic Cell- and Monocyte-attracting Chemokine-like Protein Identified by Fold Recognition Methods.
Journal of Immunology, 176, 2069-2073 (2006)
Teyra, J.; Doms, A.; Schroeder, M.; Pisabarro, M. T. SCOWLP: a web-based database for detailed characterization and visualization of protein interfaces.
BMC Bioinformatics, 7:104 (2006)
Kittler, R.; Putz, G.; Pelletier, L.; Poser, I.; Heninger, A. K.; Drechsel, D.; Fischer, S.; Konstantinova, I.; Habermann, B.; Grabner, H.; Yaspo, M. L.; Himmelbauer, H.; Korn, B.; Neugebauer, K; Pisabarro, M. T.; Buchholz, F. An endoribonuclease- prepared siRNA screen in human cells identifies genes essential for cell division.
Nature, 432, 1036-1040 (2004)
Cobos, E. S.; Pisabarro M.T., Vega, C., Lacroix E., Serrano L.; Ruiz-Sanz, J. Martinez J. C. A miniprotein scaffold used to assemble the PPII binding epitope recognized by SH3 domains.
J. Mol. Biol., 342(1):355-365 (2004)
Skelton, N.J.; Koehler, M.F.; Zobel, K.; Wong, W.L.; Yeh, S.; Pisabarro, M.T.; Yin, J.P.; Lasky, L.A.; Sidhu, S.S. Origins of PDZ domain ligand specificity. Structure determination and mutagenesis of Erbin PDZ.
J. Biol. Chem., 278(9):7645-7654 (2003)
Camenisch, G.; Pisabarro, M.T.; Sherman, D.; Kowalski, J.; Nagel, M.; Hass, P.; Gurney, A.; Bodary, S. C.; Liang, X.H.; Clark, K.R.; Beresini, M. H.; Ferrara, N.; Gerber, H.P. ANGPTL3 stimulates endothelial cell adhesion and migration via integrin αVβ3 and induces blood vessel formation in vivo.
J. Biol. Chem., 277(19):17281-17290 (2002)
Vucic, D.; Deshayes, K.; Ackerly, H.; Pisabarro, M.T.; Kadkhodayan, S.; Fairbrother, W.J.; Dixit, V.M. SMAC negatively regulates the anti-apoptotic activity of ML-IAP.
J. Biol. Chem., 277(14):12275-12279 (2002)
Wu Y.; Dowbenko D.; Pisabarro M. T.; Dillard-Telm L.; Koeppen H.; Lasky LA. PTEN2, a Golgi-associated testis-specific homologue of the PTEN tumor suppressor lipid phosphatase.
J. Biol. Chem., 276(24):21745-21753 (2001)
Skelton, N. J.; Chen, Y. M.; Dubree, N.; Quan, C.; Jackson, D.; Cochran, A.; Zobel, K.; Deshayes, K.; Baca, M.; Pisabarro, M.T.; Lowman, H. B. Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40(29):8487-8498 (2001)
Vucic, D.; Stennicke H.; Pisabarro, M.T.; Salvesen, G.; Dixit, V. M. ML-IAP, a novel inhibitor of apoptosis that is preferentially expressed in human melanomas.
Curr. Biol., 10(21):1359-1366 (2000)
Uren, A. G.; O'Rourke, K.; Aravind, L.; Pisabarro, M.T.; Seshagiri, S.; Koonin, E. V.; Dixit, V. M. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma.
Mol. Cell, 6:961-967 (2000)
Gille, H.; Kowalski, J.; Yu, L.; Chen, H.; Pisabarro, M.T.; Davis-Smyth, T.; Ferrara, N. A repressor sequence in the juxtamembrane domain of Flt-1 (VEGFR-1) constitutively inhibits vascular endothelial growth factor-dependent phosphatidylinositol 3'-kinase activation and endothelial cell migration.
EMBO J., 19(15):4064-4073 (2000)
Fuh, G.; Pisabarro, M. T.; Li, Y.; Quan, C.; Lasky, L. A.; Sidhu, S. S. Analysis of PDZ domain-ligand interactions using carboxy-terminal phage display.
J. Biol. Chem., 275(28):21486-21491 (2000)
Pisabarro, M. T. ; Serrano, L.; Wilmanns, M. Crystal structure of the Abl-SH3 domain complexed with a designed high-affinity peptide ligand. Implications for SH3-ligand interactions.
J. Mol. Biol., 281(3):513-521 (1998)
Martinez, J. C.; Pisabarro, M. T.; Serrano, L. Obligatory steps in protein folding.
Nature Struct. Biol., 5(8):721-729 (1998)
Wilcock, D., Pisabarro, M.T., Lopez-Hernandez, E., Serrano, L.; Coll, M. Structure analysis of two CheY mutants: Importance of the hydrogen-bond contribution to protein stability.
Acta Crystalographica D Biol. Crystallogr. 54(3):378-385 (1998)
Ortiz, A. R., Pastor, M., Palomer, A., Cruciani, G., Gago, F., Wade, R. C., Pisabarro M.T. Reliability of CoMFA Models: Effects of Data Scaling and Variable Selection using a Set of Human Synovial Fluid Phospholipase A2 InhibitorsPrediction of Drug Binding Affinities by Comparative Binding Energy Analysis.
J. Med. Chem., 40(25):4168 (1997) [correction from J. Med. Chem., 40(7)1136-1148]
Pisabarro, M. T. and Serrano, L. Rational design of specific high-affinity peptide ligands for the Abl-SH3 domain.
Biochemistry, 35(33):10634-10640 (1996)
Ortiz, A. R.; Pisabarro, M. T.; Gago, F.; Wade, R. C. Prediction of drug binding affinities by comparative binding energy analysis.
J. Med. Chem., 38:2681-2691 (1995)
Pisabarro, M. T.; Ortiz, A. R.; Gago, F.; Serrano, L.Molecular modeling of the interaction of polyproline-based peptides with the Abl-SH3 domain: rational modification of the interaction.
Prot. Engineering, 7(12):1455-1462 (1994)
Pisabarro, M. T.; Ortiz, A. R.; Serrano, L.; Wade, R. C. Homology modeling of the Abl-SH3 domain.
Proteins: Structure, Function and Genetics, 2:203-215 (1994)
Pisabarro, M. T.; Ortiz, A. R.; Palomer, A.; Cabré, F.; García, M. L.; Wade, R. C.; Gago, F; Mauleón, D.; Carganico, G. Rational modification of a human synovial fluid Phospholipase A2 inhibitor leading to enhanced activity.
J. Med.Chem., 37:337-341 (1994)
Pisabarro, M. T.; Palomer, A.; Ortiz, A. R.; Wade, R. C.; Gago, F.; Mauleon, D.; Garganico, G. Rational drug design: GRID- and LUDI-based structural modifications of a human synovial fluid Phospholipase A2 inhibitor leading to enhanced activity.
J. Mol. Graphics, 12(1):72 (1994)
Ortiz, A. R.; Pisabarro, M. T.; Gago, F. Molecular model of the interaction of bee venom Phospholipase A2 with Manoalide.
J. Med. Chem., 36:1866-1879 (1993)
Ortiz, A. R.; Pisabarro, M. T.; Gallego, J.; Gago, F. Implications of a consensus recognition site for phosphatidylcholine separate from the active site in cobra venom Phospholipase A2.
Biochemistry, 31(11):2887-2896 (1992)