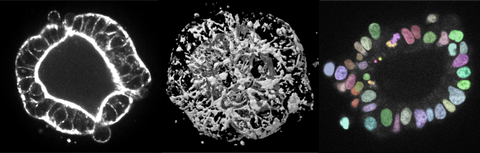
Biophysics of patient-derived cancer organoids
Rapid advancements in DNA- and RNA-sequencing technologies has made it possible to identify genetic differences between individual cancer patients. However, using such molecular-level signatures to predict overall disease progression or drug response has proven difficult. We are taking a novel biophysical approach – using an analysis of collective cell behavior and architecture in tumorous tissue to understand patient-to-patient heterogeneity in human cancers of the gastrointestinal tract. Such an analysis can provide a useful intermediate level of understanding that can help bridge the molecular and organismal scales. To do so, we are growing cancer stem cells from human patients as “organoids” – small tissues grown in 3D ex vivo culture. By acquiring a systematic, quantitative description of organoid morphology, dynamic behavior, and response to different mechanical and signaling environments, we aim to: (1) gain insight into the unknown physical mechanisms driving cancer progression, and (2) devise novel, cellular-level metrics of individual variability in drug response and disease progression.
Our previous work has taken a similar approach of analyzing the intermediate scale of biological organization – from cells to tissues – to understand tissue growth during normal development. Using the Drosophila wing for its genetic tractability and relatively simple, nearly 2D geometry, we developed tools to systematically track cellular movements, divisions, extrusions, and shape changes during multiple stages of wing development. Combined with an analysis of mechanical tissue stresses and physical models developed in collaboration with Frank Jülicher at the Max Planck Institute for the Physics of Complex Systems (MPI-PKS), we generated an unprecedented, quantitative analysis of the cellular and physical basis of tissue growth and morphogenesis. The experimental tools and theoretical concepts developed during this work form the basis for our new project studying the more complex 3D morphologies of cancer organoids.
- Connect organoid architecture and dynamics with cancer patient outcome
- Determine the biophysical mechanisms underlying variability in organoid morphology and behavior
- Design novel cellular-level metrics for predicting cancer severity and response to drug therapy in individual patients
 © Katrin Boes
© Katrin Boes
PhD Natalie Dye
|
POSITIONS |
|
|---|---|
| starting 01/2021 | Mildred-Scheel-Nachwuchszentrum (MSNZ) research group leader affiliated with Physics of Life, TU Dresden |
| 2019-2020 | Acting group leader, Max Planck Insititue of Molecular Cell Biology and Genetics (MPI-CBG), Dresden |
| 2012-2019 | Postdoctoral fellow, Max Planck Insititue of Molecular Cell Biology and Genetics (MPI-CBG), Dresden (Eaton Lab) |
| EDUCATION | |
|---|---|
| 2003-2010 |
PhD in Biochemistry, Stanford University |
| 1999-2003 | B.Sc in Biochemistry, University of Maryland |
|
HONORS |
|
|---|---|
| 2012 – 2014 | EMBO Long Term Postdoctoral Fellowship |
SELECTED PUBLICATIONS
Dye NA, Popovic M, Iyer V, Eaton S, Jülicher F. Self-organized patterning of cell morphology via mechanosensitive feedback. BioRxiv. 2020.
Dye NA, Popovic M, Spannl S, Etournay R, Kainmueller D, Ghosh S, Myers G, Jülicher F, Eaton S. Cell dynamics underlying oriented growth of the Drosophila imaginal wing disc. Development. 2017.
Etournay R, Merkel M, Popović M, Brandl H, Dye NA, Aigouy B, Salbreux G, Eaton S, Jülicher F. TissueMiner: A multiscale analysis toolkit to quantify how cellular processes create tissue dynamics. Elife. 2016
Complete publication record at ORCID
Technician
NameMay-Linn Thepkaysone
PhD student
NameSanika Jahagirdar
Send encrypted email via the SecureMail portal (for TUD external users only).
SELECTED PUBLICATIONS
Dye NA, Popovic M, Iyer V, Eaton S, Jülicher F. Self-organized patterning of cell morphology via mechanosensitive feedback. BioRxiv. 2020.
Dye NA, Popovic M, Spannl S, Etournay R, Kainmueller D, Ghosh S, Myers G, Jülicher F, Eaton S. Cell dynamics underlying oriented growth of the Drosophila imaginal wing disc. Development. 2017.
Etournay R, Merkel M, Popović M, Brandl H, Dye NA, Aigouy B, Salbreux G, Eaton S, Jülicher F. TissueMiner: A multiscale analysis toolkit to quantify how cellular processes create tissue dynamics. Elife. 2016
Complete list available at ORCID
LINKS
CONTACTS
Tba