Jan 26, 2018
Scientists from Max-Planck and Helmholtz societies develop new method for the quantitative measurement of lipid flux
Cell membranes are constantly subjected to extensive compositional remodelling in response to cellular as well as nutritional and metabolic challenges. While state of the art mass spectrometry methods would allow for the quantification of total lipid composition, the time dependent turnover of lipid species was not accessible to date. Thus, there is a need for generic and accessible analytical tools that combine full-lipidome quantification with simultaneous monitoring of the turnover, the flux, of individual lipids.
In a close collaborative work, scientists from Max-PIanck Institute for Cell Biology and Genetics (MPI-CBG), École polytechnique fédérale de Lausanne (EPFL) as well as the Institute for Pancreatic Islet Research/Paul Langerhans Institute Dresden (IPI/PLID), member of the Helmholtz Zentrum München, succeeded in the development of a shotgun ultra-high-resolution mass spectrometry (sUHR) method, which for the first time enables to quantify the absolute abundance and determine the turnover rate of lipid species by direct analysis of total lipid extracts.
“Although existing analytical technologies could quantify major lipids in one snapshot acquired at any given time, it was difficult to monitor a “concealed” lipid remodelling that does not change their overall quantities” explains Dr. Andrej Shevchenko, group leader at MPI-CBG, “therefore, we joined forces with Dr. Ünal Coskun at IPI/PLID to develop a simple and efficient method to monitor the lipid dynamics using ultra-high-resolution mass spectrometry.”
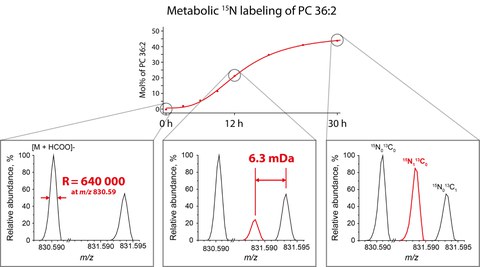
Basically, the method works as follows, cells are fed with biosynthetic precursor of common lipid classes synthesized using 15N isotope instead of 14N isotope that is common in nature. When these isotopically labelled precursors were incorporated into lipids during biosynthesis, masses of labelled lipids, in turn, increased by 1 Dalton because they contain 15N atoms instead of 14N atoms. Although this labelling process is simple and efficient, it was rarely used in analytical practice. Masses of lipids artificially labelled with 15N isotopes coincided masses of the same lipids containing 13C – a common natural isotope of carbon. Regardless of what isotope if the detected lipid happened to contain 13C or 15N isotope, its total mass differed from the mass of native (unlabelled) lipid molecule by the same 1 Dalton. However, ultra-high-resolution mass spectrometer was able to distinguish a very tiny 0,0063 Dalton difference in isotope masses and reliably detect and quantify 15N labelled lipids independently from unlabelled molecules naturally possessing 13C isotopes. Then by feeding different 15N labelled lipid precursors, the scientists were able to quantify the incorporation rates of 15N atoms into individual molecules of major lipid classes in a single analysis.
“When we think of lipids we intuitively think of dietary lipids in the context of obesity and various diseases like diabetes, or we think of the few clinical markers: cholesterol, triglycerides and lipoprotein particles” says Dr. Ünal Coskun. The lipid world, however, is more complex. Cells synthetize and turn over several thousands of different lipids. Membrane lipids are of paramount importance for the direct regulation of membrane receptors and their cellular signalling activities. In essence, cellular live is unthinkable without membrane lipids. “The new method for the first time allows us to quantitatively follow every detectable lipid and to correlate it with health and disease states. Moreover, the quantitative nature of the method is key for standardization and thus translations into clinical applications in the future.”
Original Publication:
Schuhmann et al., Monitoring Membrane Lipidome Turnover by Metabolic 15N Labeling and Shotgun Ultra-High-Resolution Orbitrap Fourier Transform Mass Spectrometry, (2017) Anal Chem. 89(23):12857-12865
http://pubs.acs.org/doi/full/10.1021/acs.analchem.7b03437