Short Tandem Repeat (STR) Analysis
Human individuals differ in the number of short tandem repeats for diverse genomic loci. These differences can be detected using multiplex PCR from genomic DNA. When analyzing a sufficient amount of STR loci the identity of a person and relations of two different individuals can be determined with high accuracy. STR profiles are thus used regularly in forensics and paternal testing.
STR profiling can also be used to identify cell lines from different passages or sources. The facility uses STR analysis to prove that a newly derived iPSC line was generated from the donor sample or that an engineered hiPSC line was derived from and is isogenic to the parental line.
Importantly, one needs a reference STR profile or sample (parental line or donor material) for comparison.
The STR profile is analyzed by multiplex PCR from genomic DNA isolated from hiPSC lines and donor samples in collaboration with the Institute of Forensic Medicine at the medical faculty of Technische Universität Dresden.
Please note that the genotype of patient samples (including hiPSCs derived from it) as revealed by STR analysis usually has to be kept confidential (in accordance with the ethics committee approval) because it might allow identification of the individual. The CRTD5 hiPSC depicted as example below was reprogrammed from commercially available BJ Fibroblasts that have a known STR profile.
The STR profile of CRTD5 hiPSCs shows the number of repeats for the depicted locus above the corresponding peak. Most loci are heterozygous - the number of repeats differs on the maternal and paternal chromosome. Three loci (D21S11, D19S433, D5S818) are homozygous with one peak only. The PCR product from the Amelogenin locus shows different sizes for X and Y chromosome, allowing a distinction between male and female lines.
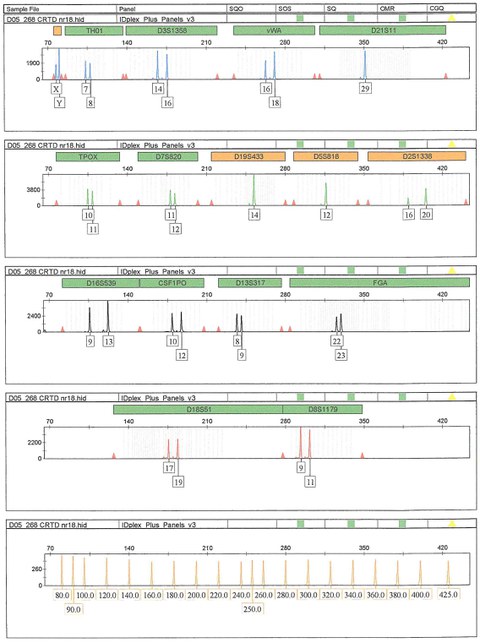
Multiplex PCR result for 16 loci with variable numbers of short tandem repeats (STR) in CRTD5 hiPSCs.
