Oct 13, 2021
New volume electron microscopy atlas of whole cells and tissues published
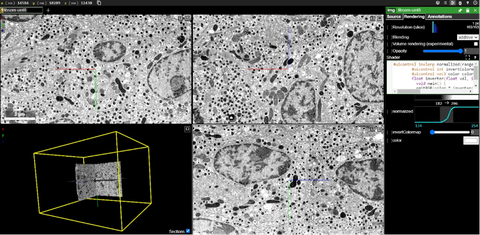
Screenshot of the https://openorganelle.janelia.org website with the xyz navigation windows and 3D view of a volume of pancreatic islet beta cells.
Understanding cellular architecture is essential for understanding biology. Electron microscopy (EM) uniquely visualizes cellular structures with nanometer resolution. However, traditional methods, such as thin-section EM or EM tomography, have limitations as they visualize only a single slice or a relatively small volume of the cell, respectively. However, focused ion beam-scanning electron microscopy (FIB-SEM) has demonstrated the ability to image larger volumes of cellular samples. Scientists of the Paul Langerhans Institute Dresden (PLID) of the Helmholtz Center Munich at the University Hospital Carl Gustav Carus and Faculty of Medicine of the TU Dresden contributed to a study lead by researchers from the Janelia Research Campus who have developed and applied a high-resolution FIB-SEM technology to establish a volume EM atlas for whole cells and tissues, including cancer cells, immune cells, mouse pancreatic islets and Drosophila neural tissues. This work has now been published in the highly renowned journal “Nature”.
Cell biologists have known the basic functions of most intracellular organelles for decades. However, despite everything scientists have learned about cellular compartments, much remains unknown about how these organelles work together. Nowadays, imaging approaches allow the gaining of insights into cells and their function, but most microscopy techniques are limited by either resolution or field of view and datasets of highly resolved whole cells are extremely rare. Researchers of the PLID have now contributed to a study which has overcome these limitations and provides for the first time large-scale high-resolution datasets of entire cells in 3D. The team lead by C. Shan Xu and Harald Hess at the Janelia Research Campus in Virginia, USA has acquired image volumes of several cell types including neurons and pancreatic beta cells at 4 nm voxel-size with a high-resolution electron microscopy technique resolving all of the cell’s organelles including even small filaments of the cytoskeleton.
“Remarkably, the acquisition of images at such resolution required the progressive and extremely accurate milling of each sample for several weeks within the microscope - an unprecedented engineering feat!” says Dr. Andreas Müller, co-author of the study, adding “This new cell atlas allows now for the first time a virtual journey inside different cell types with the possibility to observe every single organelle in incredible detail. These datasets thus provide an astonishing view into the complex organization of cells and shall help us to gain a better understanding of how they work.”
PLID scientists have already used this technology to completely reconstruct beta cells. This enabled them to gain previously impossible insights into the interaction of insulin- containing vesicles with the cytoskeleton and, for the first time, to resolve the entire microtubule network of a mammalian cell (Müller et al., 2021).
All acquired images are available at the https://openorganelle.janelia.org/ website under an open-access license. The website not only enables viewers to browse through the images. It is also possible to download the images or create shareable links of interesting features in the cell. This facilitates new findings and fosters collaboration.
Original Publication
C. Shan Xu, Song Pang, Gleb Shtengel, Andreas Müller, Alex T. Ritter, Huxley K. Hoffman, Shin-ya Takemura, Zhiyuan Lu, H. Amalia Pasolli, Nirmala Iyer, Jeeyun Chung, Davis Bennett, Aubrey V. Weigel, Tobias C. Walther, Robert V. Farese, Jr., Schuyler B. van Engelenburg, Ira Mellman, Michele Solimena, Harald F. Hess.
An open-access volume electron microscopy atlas of whole cells and tissues. Nature. Published online October 6, 2021. DOI: 10.1038/s41586-021-03992-4.
