Biomedical Genomics
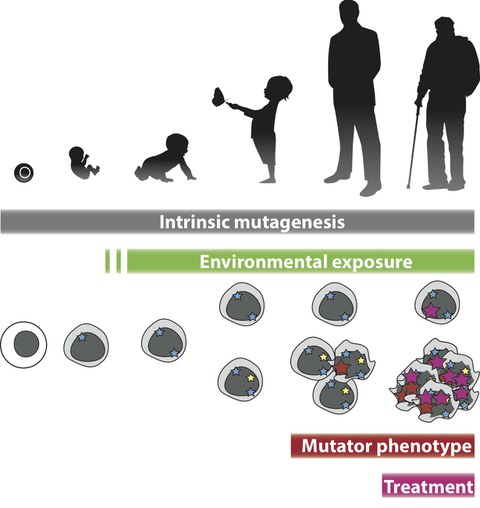
Understanding the link between genetics and the development of cancer has long been a focus of cancer research. With newest sequencing techniques, it is now possible to assess the genetic and epigenetic status as well as genome instability of individual patient’s and their individual cells at a massive scale. This can pave the way for personalized cancer therapy and may allow strategies to reduce treatment side effects on healthy cells. In individual patients, the distribution of DNA damage and mutations across the genome is highly heterogeneous. To date the mechanisms leading to individual patterns of DNA damage and somatic mutations are poorly explored. Therefore, this information is rarely used for clinical treatment decisions.
The Poetsch group employs computational techniques and machine learning approaches to assess and model DNA damage and repair processes, mutagenesis and genome editing to develop clinical applications. The group established a novel technique for genome-wide measurements of oxidative DNA damage and contributed to the understanding of genome editing precision. Work is underway to understand this in the context of different cancer types and as a consequence to different treatment regimens tissue specifically.
- Tissue-specific genome distribution of radiation induced oxidative DNA damage
- Understand how mutagenesis mechanisms shape somatic mutation landscapes in cancer
- Determinants and predictability of genome editing precision
LAB MEMBERS
 © Hagen Gebauer
© Hagen Gebauer
Group leader
NameDr. Anna Poetsch
Send encrypted email via the SecureMail portal (for TUD external users only).
|
POSITIONS |
|
|---|---|
| since 07/2020 | Mildred-Scheel-Nachwuchszentrum (MSNZ) research group leader affiliated with BIOTEC, TU Dresden |
| 2018-06/2020 | Independent scientist at St. Anna Childhood Cancer Research Institute, Vienna, Austria |
| 2015-2018 | Visting scientist at the Francis Crick Institute, London, United Kingdom |
| 2013-2018 | Postdoctoral research fellow at the Cancer Research UK/ The Francis Crick Institute/ University College London, United Kingdom with Prof. Nicholas Luscombe in Computational Biology in collaboration with Prof. Simon Boulton as a fellow of the Peter and Traudl Engelhorn Foundation |
| 2008-2012 | Research fellow of the Helmholtz International Graduate School for Cancer Researchat the German Cancer Research Center with Prof. Christoph Plass on DNA methylation in leukemias |
| 2002-2007 | Research fellow at the University Konstanz at the Japanese National Cancer Center Research Institute, Tokyo, Japan: with Prof. Mitsuko Masutani and Prof. Alexander Bürkle |
| EDUCATION | |
|---|---|
| 2008-2012 |
PhD in Cancer Biology, German Cancer Research Center/ Heidelberg University |
| 2005-2008 | Master of Science in Life Science, University of Konstanz, Germany with distinction |
| 2002-2005 | Bachelor of Science in Life Science, University of Konstanz, Germany |
|
HONORS |
|
|---|---|
| 2014 – 2016 | Fellow of the Peter and Traudl Engelhorn Foundation, The Francis Crick Institute, London, UK, University College London, UK, and Okinawa Institute of Science and Technology, Okinawa Japan |
| 2008-2011 |
Fellow of the Helmholtz International Graduate School for Cancer Research, German Cancer Research Center, Heidelberg, Germany |
| FUNDINGS | |
|---|---|
| 2020-2023 | Research grant from the Deutsche Forschungsgemeinschaft (DFG) ? |
SELECTED PUBLICATIONS
Poetsch AR. The genomics of oxidative DNA damage, repair, and resulting mutagenesis. CSBJ. (2020) Jan 7;18:207-219.
Chakrabarti AM, Henser-Brownhill T, Monserrat J, Poetsch AR*, Luscombe NM, Scaffidi P*. Target-specific predictability of CRISPR-mediated genome editing. Molecular Cell. (2019) Feb. 21; 73:1–15. * Corresponding author
Poetsch AR*, Boulton SJ, Luscombe NM. Genomic landscape of oxidative DNA damage and repair reveals regioselective protection from mutagenesis. Genome Biology. (2018) Dec. 7; 19:215. doi: 10.1186/s13059-018-1582-2. * Corresponding author
Blake SM, Stricker SH, Halavach H, Poetsch AR, Cresswell G, Kelly G, Kanu N, Marino S, Luscombe NM, Pollard SM, Behrens A. Inactivation of the ATMIN/ATM pathway protects against glioblastoma formation. Elife. (2016) Mar 17;5. pii: e08711. doi: 10.7554/eLife.08711.
for complete publication record see ORCID
 © @JakubZastapilo
© @JakubZastapilo
PhD candidate
NameMr Jakub Zastapilo
Multi-omics, DNA damage, 3D Genome, repetitive DNA
Send encrypted email via the SecureMail portal (for TUD external users only).
Visiting address:
Biotechnologisches Zentrum Tatzberg 47/49
01307 Dresden
SELECTED PUBLICATIONS
Poetsch AR. The genomics of oxidative DNA damage, repair, and resulting mutagenesis. CSBJ. (2020) Jan 7;18:207-219.
Chakrabarti AM, Henser-Brownhill T, Monserrat J, Poetsch AR*, Luscombe NM, Scaffidi P*. Target-specific predictability of CRISPR-mediated genome editing. Molecular Cell. (2019) Feb. 21; 73:1–15. * Corresponding author
Poetsch AR*, Boulton SJ, Luscombe NM. Genomic landscape of oxidative DNA damage and repair reveals regioselective protection from mutagenesis. Genome Biology. (2018) Dec. 7; 19:215. doi: 10.1186/s13059-018-1582-2. * Corresponding author
Blake SM, Stricker SH, Halavach H, Poetsch AR, Cresswell G, Kelly G, Kanu N, Marino S, Luscombe NM, Pollard SM, Behrens A. Inactivation of the ATMIN/ATM pathway protects against glioblastoma formation. Elife. (2016) Mar 17;5. pii: e08711. doi: 10.7554/eLife.08711.
LINKS
OPEN POSITIONS
Our group is looking for a postdoctoral fellow to use machine learning on functional genomics data to understand mechanisms of mutagenesis. Interested? Please get in touch: https://arpoe.github.io//arpoe.github.io/#jobs
CONTACT
 © Marc Eisele
© Marc Eisele