Redox Biochemistry
jump to: Group Publications, Research Homepage
Cellular thiol/disulfide redox networks play central roles in health as well as in disease, yet we lack the tools necessary to understand or harness their activity. The problem is one of selectivity: with so many enzymes that can engage in this type of redox reactivity, making chemical tools interface with just one network or enzyme was thought to be impossible. To tackle this, we introduced the concept of accumulating selectivity through multistep cascades: i.e. that a probe with multiple reactivity filters each focusing on different enzyme features (via topology, pKa, kinetics, thermodynamics, etc - see background) can tune out the noise to indeed select for just one target type. With chemical probes that provide precise information about the activity of these redox actors, or prodrugs that tap into disease-dysregulated activity to selectively deliver drugs, designing novel thiol redox reactivities lets us pursue multiple research avenues:
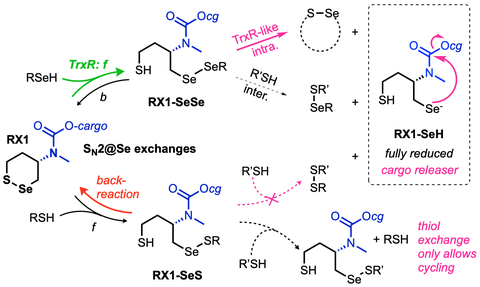
• Selenenyl sulfides (RSeSR'): from basic reactivity to cellular applications
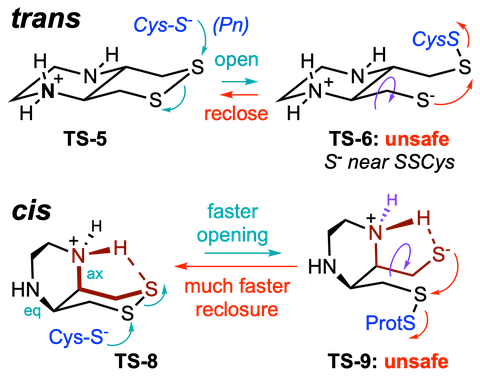
In Chem 2022 we developed the first chemoselective probes for Thioredoxin Reductase, by pioneering the deployment of cyclic selenenylsulfides. We developed scalable syntheses for these warheads in Synthesis 2023, clarified their cellular reactivity profile by quantitative in silico studies in Chem Eur J 2025, and introduced bicyclic diastereocontrol elements to improve their performance in complex environments in Angewandte 2025.
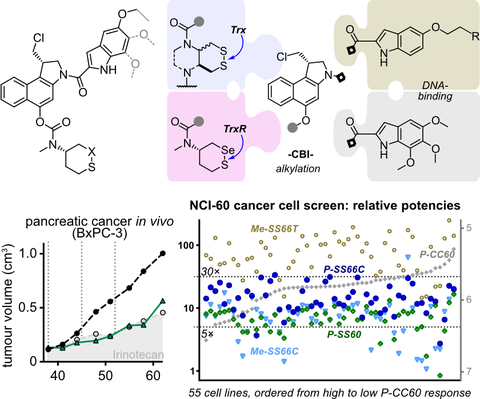
• Redox-state prodrugs and ADCs in the thiol-disulfide manifold
The clinical development of strong cytotoxins often requires increasing their tissue-selectivity of action, e.g. as we review in JACS Au 2022 for the duocarmycins. Therefore, in ACS Cent Sci 2023 we used multistep cascade designs to generate the first dithiol redoxin prodrugs for targeting the tumor microenvironment (& patent apps WO214598, WO200347, US18/587313).
• Cascade Reactivity Concepts: pioneering the links from topology to cellular redox selectivity
In JACS 2021 we introduced the principle of multistep cascades to create target-specific probes for dithiol-disulfide oxidoreductases, by cumulating layers of selectivity through kinetic tuning. We then identified the rate-limiting steps for their performance, and in JACS 2024 we could introduce the first cellularly-effective thioredoxin-targeting probes and prodrugs. In Helv Chim Acta 2025 we instead develop an irreversible dithiol-selective alkylator by applying similar cascade principles ("reversible step 1, selectivity step 2"), again installed through topology, molecularity, and reaction bias, now to elucidate cell-surface uptake mechanisms.
• Stringent biochemical evaluations for high quality redox probes
in Nat Comms 2022 we re-evaluate the last decade's papers on cyclic 5-membered disulfides. We show that kinetic lability drives unspecific effects and invalidates all prior conclusions from cellular tests.
We showed why misinterpretations arose, and presented a robust design scheme and evaluation system that promote stringent standards for redox research.
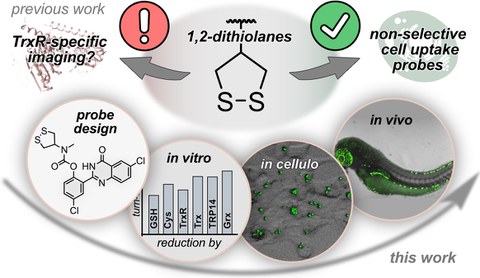
• Thiol-mediated cellular uptake and reversible, cyclic topology thiolate traps
That thiols exposed at the cell surface can 'sponge up' reversibly-alkylating species, vastly increasing the rates of cellular uptake of even gigantic cargos (viruses, nucleic acids), is now supported by a large body of evidence - much of it collected and systematised by Stefan Matile (Uni Geneva). We are interested in clarifying the molecular mechanisms underlying these TMU effects, e.g. by adding our cyclic selenenyl sulfide reagents (Chem Eur 2024) and biochemical redox target hypotheses (JACS Au 2023) into the field, and in particular by testing the hitherto cryptic role of vicinal-dithiol proteins in these processes (Helv Chem Acta 2025).
Extended Background
Cellular networks of redox reactions are crucial to driving and regulating cellular physiology, with key roles from metabolism to signaling, protein activity regulation, and gene transcription. These networks are also significantly deregulated in serious pathologies including cancer and inflammatory diseases. These redox networks tend to rely on a few key upstream players, from which reducing/oxidising equivalents are distributed to a much broader range of downstream substrates.
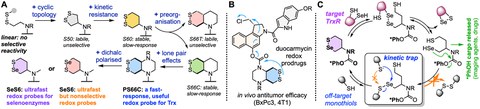
Therefore, developing selective chemical tools to respond to the activity of these key redox players is of particular interest to understand their physiological dynamics, as well as to respond to their pathological deregulation. We mainly focus on the thioredoxin (Trx) network, with some work also towards other disulfide/dithiol-manifold systems such as the Grxs, GPxs, and PDIs. Developing small-molecule selective tools for these enzymes is a highly challenging prospect, because their chemistries are so similar, and because their activity and reactivity is strongly controlled by compartmentalisation and redox fluxes. Before our work, there were no validated chemical tools for these disulfide/dithiol-manifold species that are enzyme-selective in the cellular context. Our goal is to introduce layers of selectivity by stepwise kinetic and thermodynamic tuning (A), and by rational mechanism-based design of reaction cascades (C), to tap the potential of redox-responsive small molecules, towards high-selectivity probes of redox physiology, and to image and treat redox-dysregulated diseases (B).
Our redox research is therefore pioneering novel redox-responsive chemotypes that can be used in modular "trigger-cargo" constructs to activate imaging agents or drugs conditional on the activity of specific redox enzymes and proteins (A). In parallel, we are promoting systematic methods to assess redox selectivity in the cellular context, revealing the complexities that have blocked previous tool designs from achieving meaningful impacts in redox research. Our research programme includes novel chemotype development, their use for fluorogenic imaging probes, then their adaptation into toxogenic prodrugs for cellular testing, and lastly the adaptation of selected agents into in vivo anticancer prodrugs (B).
For our full list of papers on redox reagent development and applications, see the Publications page.