SelWineQ - Prädikive Züchtung auf Weinqualität
Das Ziel von „SelWineQ“ ist es, robuste und geeignete Modelle zur Vorhersage des genetischen Qualitätspotenzials von Weinrebensorten zu entwickeln.
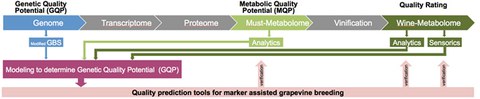
Für die Modellierung und eine Vorhersage des genetischen Qualitätspotentials anhand von Genomdaten werden geeignete phänotypische und genotypische Merkmale identifiziert. Diese Qualitätsmarker werden anschließend auf das Züchtungsmaterial angewendet. Durch dieses Vorgehen kann die Effektivität des Züchtungsprozesses drastisch erhöht werden, da voraussichtlich schlechte Genotypen durch negative Selektion frühzeitig ausgeschlossen werden. Die Erschließung und Charakterisierung von genetischen Qualitätsmarkern zur Weinqualität erfolgt über ein “Modified Genotyping by Sequencing” (GBS) Verfahren. Da das Genom von Vitis vinifera mindestens zu 70 % aus repetitiven Sequenzen besteht, kann nur ein geringer Teil der Sequenzdaten zur Identifizierung solcher Marker genutzt werden. Durch das Entfernen repetitiver Sequenzen vor der Sequenzierung der Genotypen können somit Kosten und Aufwand für die Datenanalyse im großen Maße eingespart werden.
Die Vorhersage des genetischen Qualitätspotentials von Weinreben, basierend auf dem Genotyp bis hin zum Metabolotyp und Sensotyp, kann den Züchtungsprozess um bis zu 10 Jahre beschleunigen und es entstehen neue Rebsorten für einen nachhaltigen Weinbau.
Kontakt:
Prof. Dr. Stefan Wanke (E-Mail)
Dr. Lena Frenzke (E-Mail)
Dr. Torsten Wenke (E-Mail)
Kooperationspartner:
Julius Kühn-Institut, Bundesforschungsinstitut für Kulturpflanzen
asgen GmbH & Co. KG
Rheinlandpfalz – Dienstleistungszentrum ländlicher Raum
meta-sys-x
Förderung:
Bundesministerium für Bildung und Forschung
PLANT 2030