Nov 08, 2023
Farewell to Single Well: B CUBE Researchers Automate a Popular Biophysics Technique
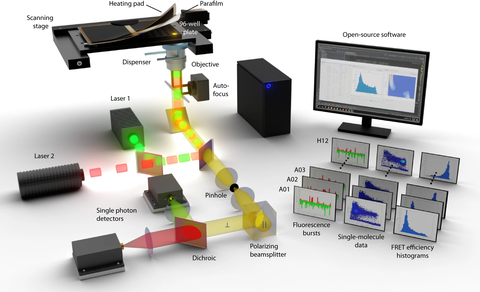
Illustration of the novel fully automated smFRET setup for high-throughput single molecule experiments. The authors provide detailed guide for building or adapting existing smFRET setups as well as an extensive open-source software suite for data acquisition, processing, analysis, and visualization.
Biophysics uses physics to advance our understanding of life from the cellular to the molecular scale. It has revolutionized biology in recent years. Now, the automation of a powerful technique in the biophysicist’s toolbox might open new avenues for biophysics. Scientists from the Schlierf group at the B CUBE – Center for Molecular Bioengineering (TUD Dresden University of Technology) presented a fully automated smFRET instrument. In a new publication published in the journal Nature Communications, the group provides detailed information on the setup, an open-source software suite for measurement, data analysis, and data visualization, and shows the possibilities of the new system using a variety of experiments with increasing complexity up to drug screening for protein misfolding.
Single-molecule Förster Resonance Energy Transfer is a powerful technique used by biophysicists to determine the structure, dynamics, and function of molecules of life - mostly proteins, nucleic acids, and lipids. It is used to determine if two points are within a certain distance of each other. smFRET has a wide variety of applications some of which include drug-protein interactions, DNA repair, recombination, and replication, or protein folding.
“smFRET measurements are at a nanometer scale. Experiments are susceptible to various influences and typically require many conditions: different solutions, different concentrations of molecules, not to mention standard experimental replicates,” says Dr. Andreas Hartmann from the Schlierf group at the B CUBE. In reality, a typical smFRET experiment often requires rapidly over 50 experimental conditions and this means days or weeks of manual work spent in the laboratory.
“Working with smFRET for over 10 years now, we’ve identified the need for automation. So far, no company could offer such an automated system. This is why we set out to build one ourselves,” says Prof. Michael Schlierf, research group leader at the B CUBE.
The team determined a set of prerequisites. First and foremost, the automation couldn’t compromise the precision and accuracy of the measurements. The system had to offer a chance to measure many conditions at once. It had to be easy to build and, at the same time, easy to integrate into existing smFRET setups. Finally, the software used for operating the instrument had to be user-friendly and open-source, enabling data analysis without expert knowledge.
Multiwell Bliss
The new system is fully automated and allows measurement of up to 96 different experimental conditions, potentially even up to 384 in the future. The system is based on multiwell plates that are commonly used in laboratories. In such a way, one can set up an experiment with all the necessary conditions and varieties in one go and leave the machine to run unattended for 32 hours or more, before getting ready to go through the results that are analyzed automatically on-the-fly.
“Our system frees massive amounts of hands-on time for scientists. Not only do the experiments run unattended but being able to set up 50, 60, or 90 experimental conditions to run at the same time saves weeks of work. These are weeks that can be already used on interpreting the data, coming up with, and testing new hypotheses,” explains Dr. Hartmann.
Precision Measured in Ångströms
The team tested the new system in a thorough series of proof-of-concept experiments, with each of the experiments increasing in complexity and – in turn – increasing its demands on the system.
First, the team employed a so-called DNA nanoruler to measure the accuracy and precision of the system. “It’s a clever and predictable tool that emerged in recent years as a way to quantify the performance of smFRET instruments,” explains Dr. Hartmann. “We could show that our automated system does not compromise on the precision and accuracy. We could discriminate between the wells in the multiwell plate on an Ångström scale, i.e., a hundred-millionth of a centimeter.”
In a variety of experiments, the team could show that the system can be successfully used to study DNA nanostructures, protein folding, protein-protein competition, and drug-protein interactions.
Accessible to Everyone
This proof-of-concept study goes one step further and offers ready-to-use guides for setting up the system in a lab. “Since smFRET is a technique that is widely used, it was vital for us to offer everyone a way of adapting their existing setups to make it automatic. All the components of the system are commercially available and can be installed without expert knowledge,” says Prof. Schlierf.
To bring the system to everyone, the team also provides a comprehensive software suite focusing on every step of the process, from data acquisition through processing, analysis, and finally, visualization. The software is open-source and provides an extensive tutorial, which allows everyone to integrate the code into their existing workflows.
“With the automation of many different methods in the biology and biophysics fields, it was a matter of time until smFRET became less hands-on. We are happy that we could contribute to making this happen. We hope that this will unload a lot of time and resources in the labs around the world. It certainly did in ours,” concludes Prof. Schlierf.
Original Publication
Andreas Hartmann, Koushik Sreenivasa, Mathias Schenkel, Neharika Chamachi, Philipp Schake, Georg Krainer and Michael Schlierf: An automated single-molecule FRET platform for high-content, multiwell plate screening of biomolecular conformations and dynamics. Nature Communications (October 2023)
Link: https://doi.org/10.1038/s41467-023-42232-3
Github Link: https://github.com/SchlierfLAB/autoFRET
