Biomedical Genomics
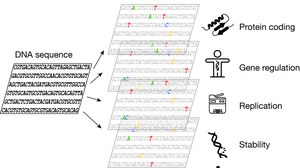
We study how the genome encodes different layers of information, specifically its own stability, repair, and mutagenesis. Using a combination of computational biology, machine learning, deep learning models for sequence data, clinical and experimental data from collaborating labs, we try to understand the underlying mechanisms despite their complexity, and look for routes to use this knowledge to understand the biology of ageing and cancer development.
In 2026 the lab will move to CECAD and the Institute for Genetics in Cologne.
 © Anna Poetsch
© Anna Poetsch
Our research
The Biomedical Genomics group employs computational techniques, machine learning, and deep learning approaches to assess the layers of information genomes encode. In this context we are particularly interested how the genome functions in the context of balancing function with DNA damage and mutagenesis.
The group established a novel technique for genome-wide measurements of oxidative DNA damage and contributed to the understanding of genome editing precision and the genome wide distribution of mutations.
For this we use computational biology with different types of functional genomics data (AP-Seq, HiC, ChIP-Seq,...) and cancer genomics data and different machine learning and deep learning techniques, particularly the DNA language models we develop, GROVER and EAGLE-MUT.