Gene Targeting Guide
The content of this page is not getting updated any longer. For the newest developments in genome editing please see our recent publications. The methods listed here are still of use and supply a general understanding of design for gene targeting constructs.
We describe a way to design conditional alleles for mouse as well as practical hints for creating them. However, the strategies described in this page can be transfered to other applications.
"For a knock-out we want to cause the most disruption to the protein, with causing the least disruption to the gene (genome)."
The creation of the constructs is based on recombineering. Please, see our Recombineering Guide for information about this technology.
This guide is based on the ENSEMBL Release 59 (August 2010).
Last changes made to the content: Feb 2012
This Gene Targeting Guide for knock-out first allele constructs is similar to the following strategies used by different consortia:
For a strategy overview of our Gene Targeting pipeline, please see section 15)
Look up your gene of interest
in ENSEMBL (and UCSC)
Example: Ctr9 ( ENSMUSG00000005609)
- note down gene names, accesion number and other gene identifiers (location, protein features)
- look up whether there is already a Knock--Out Cell Line or Targeting Vector available at International Mouse Phenotyping Consortium (IMPC)
| Information | Example | |
|---|---|---|
| strand? | forward | pic1 |
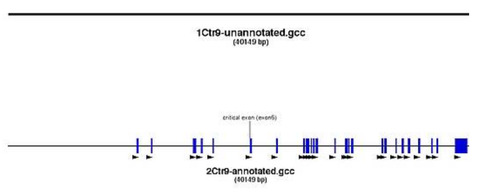
| different transcripts? →usually go for the longest one (note down acc. #) | 2, differ at the later exons | pic1 |
| exons? | several (more than 2!) | pic2 |
| Havanna Genes (Location view → configure this page → genes → display Havanna genes) | Ctr9 longest transcript is Havanna confirmed | |
| existing KO alleles (configure this page → external data → Ko alleles) | 2 KO alleles for Ctr9 in the pipeline (to learn more about the projects right click on it, you might consider ordering the targeted ES cells or the targeting vector from EUCOMM) | pic3 |
| existing KO designs (manage your data → attach DAS → Sanger → check KO_designs) | Many KO designs existing for Ctr9 | pic3 |
| UCSC | ENSEMBL information confirmed | pic4 |
| strain variation for that gene (SNP, etc.) | nearly no differences between B6 and 129 |
Is the gene of interest expressed in ES-cells?
- check with the International Gene Trap Consortium (IGTC) for existing cell lines
- expressed genesd are suitable for promoterless targeting cassettes
Example: Ctr9 has 2 different gene traps (pic)
Selection of a target exon for a knock-out construct
- view exons of the transcript you have chosen as design basis (pic)
- look for the most 5' exon that meets the following criteria
| Exon selection criteria and example for Ctr9 | |||||||
|---|---|---|---|---|---|---|---|
| Reason | 1 | 2 | 3 | 4 | 5 | 6 | |
| frame shifting exon? (exon phases) | deletion will lead to early stop codon | no | no | yes | no | yes | |
| exon present in all (translated) transcripts? | otherwise possibility of function replacement by other protein product | yes | yes | ||||
| exon in first half of coding sequence | otherwise function might not get affected | yes | yes | ||||
| (exon coding for domain?)** | esp. impotant if no frameshifting exon is available | yes | yes | ||||
| flanking introns bigger than 500 bp [150 bp]?* | to leave splice acceptors and donors intact | no | yes | ||||
| exon smaller than 2 kb | increasing distance of loxP-sites up and downstream of the exon, decreases the efficiency of recombination between them | yes | |||||
| (exon bigger than ~50 nt?)** | very close SSR sites do not recombine well, and very small deletions might be without effect | yes | |||||
| 5 kb upstream of the exon does not contain the gene's promoter region? | for promoterless targeting | yes | |||||
| (at least 35 aa of the protein are made in the KO)** | otherwise potential to re-initiate translation | yes | |||||
| no antisense genes in this region | otherwise affecting another gene by KO | yes | |||||
| (at least 55 bp from the PTC to the last splicing donor)** | nonsense mediated decay (NMD) of tghe RNA is prohibited, so mRNA levels might be detectable | yes | |||||
* 500 bp is the minimum size of flanking introns in the EUCOMM designs, 150 bp is usually enough to put a cassette without interruption of splice adaptors and splice donors (leave at least 50 bp from the splice donor, which is 5' end of the intron, and 100 bp from the splice acceptor, which is the 3' end of the intron)
**those criteria are taken in account by EUCOMM designers and are not the most important for us
- note down exon identifier and position
Exceptions:
- If flanking introns are too small or the exon itself is very small, try to knock out 2 sequential exons (check for frame shift again)
- For one intron genes, put the downstream loxP-site into the 3' non-coding region or downstream of the polyadenylation signal
Examination of the critical exon's environment
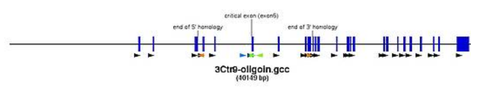
- view chosen exon and at least 5 kb up and downstream in Location view (pic)
- check for regulatory elements and repeats in this region
| Element | Reason | How to view | Ctr9 |
|---|---|---|---|
| CpG-islands | placing cassettes into CpG island can lead to silencing of the selection markers or altered expression of the gene | Ensembl Location View -> Configure this page -> simple features -> CpG-islands | won't be hit by he cassettes |
| Conservation | conserved regions are a hint to regulatory elements that you do not want to hit with a cassette due to possible pleiotropic effects | Ensembl -> Configure this page -> Multiple alignments -> constrained elements | only at the exons, so won't be hit by the cassettes |
| Repeats | repetitive sequence decreases the correct recombineering proportion when placing the cassettes or subcloning from the BAC, and repetetive regions at the homology arm ends might decrease the homologous recombination efficiency in ES cells | Ensembl -> Configure this page -> Repeats -> all repeats | enough distance from critical exon to put cassettes, homology arm end region also repeat-free |
| HCNE (highly conserved nonconding elements) | see conservation | Ancora browser -> give position and enable all HCNEs | none (pic) |
- to define the positions of the cassettes as well as the homology arm length (by subcloning), you have to select oligos of 50 bp [70 bp]* which serve as homology arms in recombineering
- to design those oligos you can use the
- recheck also the G-oligos for not recombining in exons (if small deletions happen) and in CpG-island (chromosome structure might inhibit recombination)
HTGT-tool
- create an account under http://www.sanger.ac.uk/htgt
- use custom design tool
- create a design for knock out first, block specified (means not fixed to specific bp)
- set parameters (taking intron sizes, repeats, HCNEs, ... in account)
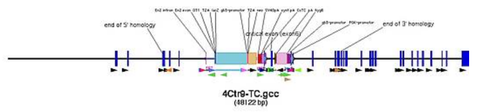
changes of default for Ctr9 (pic)
homology arm length: 5000 bp
upstream spacer: 250 bp
- insert Ensembl ID: ENSMUSG00000005609
- select critical exon(s): ENSMUSE00000203310
- change comment line (otherwise your email address is visible with the designs in ENSEMBL)
- Create&Run
- note down ID
- click refresh or check for design by ID later (pic)
- in case design failed or you want to change it, click Edit and change parameters as described above (you may delete the old design then)
Manual Design
- neccessary for non-mouse DNA, non-exon targeting or non-annotated genes
- download gene sequence into your vector construction program
- select the positions in the same distance from the critical exon as for the HTGT-tool
- blast about 150 bp to the genome
- find non-repetetive regions (exclude low complexity sequence)
- check for around 50% GC content
- (exclude hairpin formation etc. if software available)
- decide on 50 bp* serving as oligo (homology arm for recombineering)
*70 bp should be used for subcloning oligos.
Retrieving the sequence
- click on critical exon to get it in the Ensembl location view
- export data, add 20,000 bp flanking sequence on either side (necc. for southern design)
- export as genbank file if you vector program has an import function for that
- otherwise export as text and manually annotate
- in case you used the HTGT tool you can also copy and paste the 15_E_15 sequence, which is exon plus 15 kb up and downstream sequence
Annotation of the oligos in your file
- mark the oligos in the genomic sequence
- see where homology arms will end and which small intronic regions will be deleted by the cassette insertion (keep in mind for southern strategy)
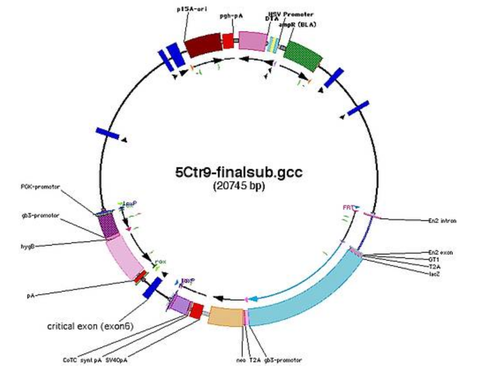
Creation of the targeted allele
- create a new file and place your cassettes
- 1st cassette here: FRT-SA-GT1-T2A-lacZneo-CotC-FRT-loxP
- 2nd cassette here: rox-PGK-HYG-rox-loxP
- ATTENTION: always check the orientation of the loxP-sites as different cassettes bring loxP-sites in different orientations. Only loxP sites that point in the same direction will give the intended deletions.
- early southern strategy design is recommended (certain designs may have no good possibilities for southern probes or enzymes and therefore have to be altered, best before practical work starts)
- design the southern strategy by comparing wildtype allele to targeted allele
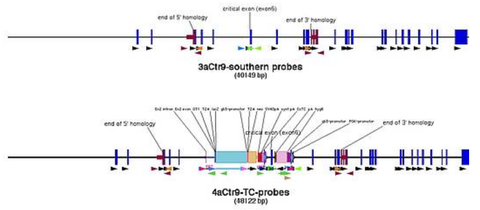
Finding southern probes
- blast about 1000 bp left of 5' homology end and right of 3' homology end to the mouse genome (set blast to "no filter")
- find at least 500 bp continuous sequence without repeats and create primers in it, that give a PCR product of 500-700 bp (good southern probe size)
- the closer the probe is next to the ends of the homology, the bigger is the chance to find good enzymes
- probes that stretch over exons are favourable
Finding restriction enzymes
that cut neither in the homology arms nor in the southern probes, meaning for the 5' southern you pick enzymes that do not cut in the region between the 5' fwd primer of the probe and oligo U5, and for the 3' southern enzymes that do not cut between oligo D3 and 3' rev primer of the probe (pic)
that are not inhibited by eucaryotic methylation patterns (table)
that give a band (when hybridized with the labelled probe) of 5 to 15 kb
that give a size difference between WT and targeted allele of > 1kb
- here: for 5' ScaI gives a band at 9888 bp for WT and 11881 bp for the targeted allele
- have at least one alternative in case enzyme cuts unexpectedly
- you may do double digests if no simple digest meets the criteria (check for enzyme buffer compatibility)
- create a theoretical gel picture for the outcome of the southern showing the size of the signal after developing the membrane (pic)
- repeat all steps for the 3' southern
If you cannot find nice probe regions or good enzymes, try to redesign the homology arms.
Creating the targeting vector in silico
- copy the sequence between the homology arm ends from the targeted allele and paste it to the backbone
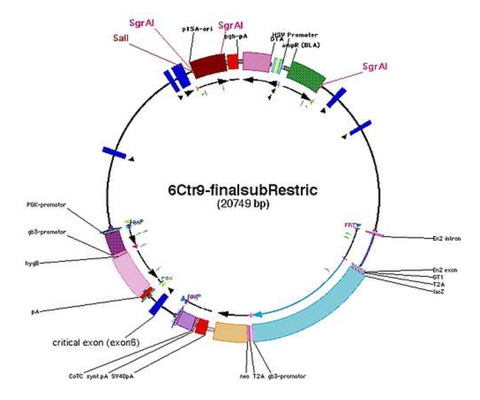
Linearisation Sites
- check for restriction sites in the backbone for linearisation
- use (combination of) restrictions sites that cut out backbone (example: SgrAI)
- or insert a new restriction site into junction between homology arm and backbone on both sides (for using the counter selection marker on the backbone [TK/DTA] only on one side)
- example for Ctr9: SalI (GTCGAC) or SnaBI (TACGTA)
- note down on which side the counter selection marker will remain after digestions
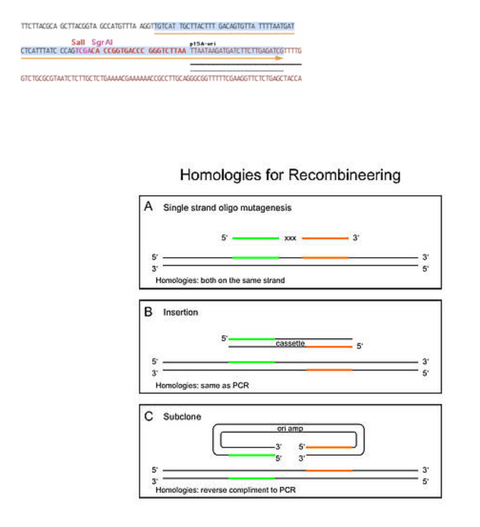
Oligos for ordering
- get the complete sequence of the oligo by copying the 50/70 bp homology (designed by HTGT) plus the primer for cassette/backbone amplification
- [please note that for subcloning by gap repair (see C in figure below), the sequences included in the oligonucleotides should be the inverse complement to the orientations which are for insertions (see B in figure below)]
- by copying it out of the final file, there is no chance of miscombining the 2 parts
- do not forget to make the reverse complement of the downstream primer
Important information
→ Overview of mouse and human BAC libraries including informations about DNA origin, backbone and selection markers.
Finding a BAC and ordering it
- we recommend to use UCSC to find BACs, as they only show end-sequenced BACs and have prooven by experience to be more reliable in the annotation (we rarely see BACs with the same name, having different length in the 2 browsers, especially for human)
- to display the BACs:
| BAC Library | Genome Browser | How to Display | Supplier |
|---|---|---|---|
| RP23/RP24 (mouse) |
ENSEMBL | Location view -> Configure this page -> External data -> switch on BAC map | CHORI BacPac |
| UCSC | switch on BAC End Pairs | ||
| CH29 (mouse) | ENSEMBL | Location view -> Configure this page -> Other DNA Alignments -> switch on CHORI-29 clones | |
| bMQ/129 (mouse) | ENSEMBL | Location view -> Configure this page -> Other DNA Alignments -> switch on ens_m37_129AB22 | SourceBioscience (Geneservice) |
| RP11 (human) |
UCSC | switch on BAC End Pairs | CHORI BacPac |
| ENSEMBL | Location view -> Region overview | CHORI BacPac |
|
| Location view -> Region in detail -> Configure this page -> misc features -> switch on 32 k clone set (you may try using a different browser than Firefox) | |||
| CH17 (human, w/o loxP sites) | UCSC | follow the instructions by CHORI to add the CH17 track to UCSC | |
| other libraries | UCSC | check for instructions by CHORI to add your favourite library to the browser |
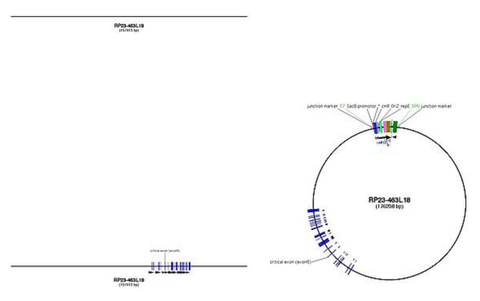
Example for Ctr9: RP23-463L18 (118,072,655-118,240,067)
There is also a clone finder at NCBI, that reads out many more libraries. We have no experience with its completeness and reliabily though so far.
Features for a good BAC
- BAC needs to contain at least the critical exon plus 5 kb up and downstream
- better: find a BAC that contains the whole gene plus 5'UTR, so that it permits usage for BAC transgenesis projects as well as for targeting issues
- the BAC should not be too big, better go for 150 kb than 300 kb, because internal rearrangements are less likely
- search for the BAC in NCBI and take one that is at least sequence confirmed on one side (SP6 or T7), better on both (use the UCSC browser to find only BACs with end-sequencing information, use the direct link from the BAC to NCBI to look up the end-sequencing information)
Creating the BAC map
- when you have found the best BAC, right click on it and get its end points
- export the sequence between this endpoints (for Ensembl: location view -> export data -> no flanking sequence)
- this sequence is the insert sequence
- if you want information about the BAC library backbones, see table or visit CHORI website
- put the sequence into your DNA depiction software and annotate
- create all intermediate files from the WT BAC file to final subclone according to the actual strategy in the lab
- use these files to decide on enzymes for restriction analysis for the intermediate steps (detection of deletions or mixtures!)
For ssOligo-experiments and backbone modifications (e.g. PiggyBac integration)
-> Overview of backbone maps and replication orientation.
What needs to be genotyped in the modified ES-cells/mouse?
- checking for the presence of the 3' loxP (especially if it is a single loxP without selection marker)
- checking for successfully Flped allele (loss of lacZneo-cassette)
- checking for successfully knocked out allele (loss of critical exon)
Where to design primers?
- put primers into WT sequence and distinguish the WT from the targeted by size difference - in case of a heterozygous gene (+/-, +/F, ...) you will always get both bands
- in case of low size difference or huge distance of primers, put an additional primer into the cassette/loxP and prove the existence of the cassette/loxP by getting a product
make the primer longer than 21 nt and have 3 non-matching nt at the end with a C/G at the very 3' end
| trapping cassettes | FRT-SA-IRES-lacZneo-pA-FRT-loxP | reading frame independent trapping cassette (description see blow) |
|---|---|---|
| loxP-FRT-SA-IRES-lacZneo-pA-FRT | same as above with different loxP position | |
| FRT-SA-IRES-lacZBSD-pA-FRT-loxP | reading frame independent trapping cassette with different selection marker for second allele targeting | |
| FRT-SA-GT0/1/2-T2A-lacZneo-pA-CoTC-FRT-loxP | reading frame specific versions of the trapping cassette with T2A (description see below) | |
| FRT-SA-IRES-BSD-pA-FRT-loxP | reading frame independet trapping cassette with different selection marker for second allele (for constitutive knockout by deletion of critical exon) | |
| FRT-SA-IRES-lacZneo-PGKBSD-pA-FRT-loxP | reading frame independent trapping cassette with additional promotordriven selection | |
| 3'loxP insertion cassettes |
loxP-PGKneo-loxP | after Cre expression single loxP remains |
| loxP-zeo-loxP | after Cre expression single loxP remains | |
| cm-loxP | after digestions and religation single loxP remains | |
| rox-PGK-Em7-BSD-rox-loxP | vehicle for 3' loxP (description see below) | |
| rox-PGK-Em7-HYG-rox-loxP | vehicle for 3' loxP (description see below, alternative selection marker) | |
| backbones | pBR322-amp | middle copy minimal vector |
| p15A-HSVtk-TK-amp | middle copy vector with bacterial selection marker and eucaryotic counter selection marker | |
| p15A-HSVtk-DTA-amp | middle copy vector with bacterial selection marker and eucaryotic counter selection marker |
Detailed description of the most important cassettes:
FRT-SA-IRES-lacZneo-pA-FRT-loxP (Chen and Soriano, 2003)
This stop cassette includes the Engrailed-2 splice acceptor to capture the transcript of the target gene. It is followed by an internal ribosome entry site (IRES) promoting the expression of a b-galactosidase-neomycin resistance fusion protein and a 3'-non-coding region ending in a polyadenylation signal. The neomycin resistance gene has an additional bacterial promotor (in frame). The cassette is flanked by FRT sites, allowing to restore gene function after the trapping by its removal via Flp expression. The loxP-site is unaffected by this recombination event.
FRT-SA-GT0/1/2-T2A-lacZneo-pA-CoTC-FRT-loxP
This cassette is the new version of the previous trapping cassette. The splice acceptor has been shortened and the IRES has been replaced by a 2A ribosomal-error peptide (Szymczak et al, 2004). This makes the cassette smalle but the absence of an IRES requires the cassette to be in frame with the exon upstream of it. Consequently the stop cassette has been made in three versions. During design, the correct version needs to be selected according to the 0,1 or 2 reading frame of the upstream exon. A second 2A peptide is included 3' of b-galactosidase, so that it is a separate protein from neomycin resistance, which enhances performance of both proteins compared to the fused version. In addition to the poly adenylation signal the cassette has a co-transcriptional cleavage site. The position of the FRT and loxP sites is as described.
rox-BSD-Em7-PGK-rox-loxP
This cassette introduces the 3?loxP site into the 3?intron. It includes a PGK promoter driving the eucaryotic and an Em7 promotor driving the bacterial expression of the blasticidin resistance gene. For genes that are expressed in ES cells and can be targeted by selection of neomycin/G418 resistance driven from the endogenous target gene promoter (termed ?promoterless selection?), the rox cassette should be removed in E.coli by expressing Dre recombinase from pSC101-BAD-Dre (Anastassiadis et al, 2009) to leave the 3?loxP site in the targeting construct. However, in a few cases after targeting in ES cells, we have found that all targeted clones omit the 3?loxP site. In these cases, the targeting can be repeated using the construct including the rox cassette and selecting for both neomycin and blasticidin resistance to force the inclusion of the 3?loxP site. For genes that are not expressed in ES cells, the targeting construct must include a promoter to drive expression of the selectable gene. Here the rox cassette provides this function. In the cases when the rox cassette remains after targeting, it must be removed either by expressing Dre in ES cells or by crossing to a Dre deleter (Anastassiadis et al, 2009).
p15A-HSVtk-DTA-amp
The subcloning vector, p15A-HSKtk-DTA-ampR, is based on the p15A plasmid origin. Consequently it does not give the very high copy yields delivered by the mutated colE1 origins in pUC-type plasmids. We have found that these unnaturally very high copy plasmids can provoke recombineering problems, so prefer p15A or the unmutated colE1 origin from pBR322 for recombineering applications, particularly subcloning by gap repair. In p15A-pTK-DTA-ampR, the diptheria toxin A chain (DTA) coding region under the HSVtk promoter lies between the p15A origin and the ampicillin resistance gene. For subcloning, the vector is amplified by PCR to attach the homology arms. The presence of a DTA gene permits the use of positive/negative selection (Mansour et al, 1988; Yagi et al, 1990). We have found that negative selection can benefit promoter-driven selection but has little effect on promoterless selection. Therefore we recommend that the vector be entirely cut off for promoterless selection or linearized for promoter-driven selection.
| Category | Link | Purpose |
|---|---|---|
| Genome Browser |
ENSEMBL | basis for designs |
| UCSC | additional information to ENSEMBL | |
| Ancora | HCNE browser | |
| ENSEMBL (old version) | based on NCBI m36 mouse assembly, for reference to old projects | |
| VEGA | ||
| Genome Engineering Databases |
International Gene Trap Consortium | |
| International Knockout Mouse Consortium | ||
| EUCOMM (European Conditional Mouse Mutagenesis Program) | ||
| BLAST tools |
Blast 2 sequences (NCBI) | |
| Megablast (NCBI) | ||
| Blast against E.coli genome | ||
| Blast against human genome (NCBI) | ||
| Blast against mouse genomce (NCBI) | ||
| BLAT (Ensembl) | ||
| Genomic tools | High Troughput Gene Targeting (HTGT) tool (SANGER) | Oligo design for targeting attempts |
| DNA tools |
PlasmidMapper | Annotation help for fasta sequences |
| In-Silico Primer Check (UCSC) | Check primers for additional unwanted products when used with genomic DNA | |
| Primer Calculator (Sigma) | Primer Analysis for T, structures, dimers | |
| Primer 3 | Primer Design for PCRs | |
| Enzyme tools |
Enzyme finder (NEB) | for availability and features of enzymes |
| Double digest finder (NEB) | to check for condition compatibility of 2 enzymes | |
| Rebase | alternative enzyme search | |
| BAC Suppliers | CHORI BacPac | |
| Geneservice | ||
| Oligo Supplier | Biomers | used by Stewart lab, any other supplier might be fine |
| DNA prep Supplier | Invitek Molecular | usage see practical protocol |
| Course about Recombineering and Mouse Genome Engineering | SANGER Wellcome Trust Advanced Courses | Genetic Manipulation of ES cells |
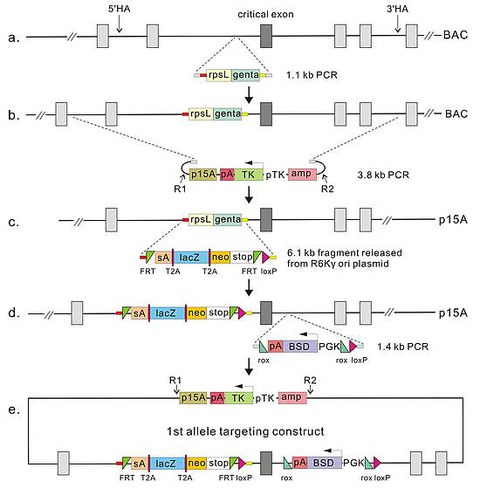
Gene Targeting Pipeline for Standard Conditional Alleles
PCReactions and Cassette Preparation
Three PCR products are required to generate the first allele targeting construct.
The first PCR product is amplified from the rpsL-gentamycin selectable/counterselectable cassette. The PCR amplified rpsL-gene cassette already contains the homology arms for the stop cassette (step 3). Consequently incorporation of the rpsL-gene cassette into the BAC by selection for gentamycin resistance incorporates the homology arms for the stop cassette. We include this step because it avoids PCR amplification of the 6.1kb stop cassette. Instead a restriction fragment from a pR6K-amp-lacZneo plasmid preparation is used, thereby reducing unwanted, PCR-based, mutagenesis.
The second PCR product is the amplification of the subcloning vector, p15A-pTK-DTA-ampR.
The third PCR product is amplified from the pR6K-PGK-BSD template, which will insert a loxP site on the 3' side of the chosen exon.
| rpsL-gen 5' | GCGTGTTTCGAGCATGTTTCTGCGTAGTGTCAGCTCATC* |
| rpsL-gen 3' | GTGTCCATCATCCTGTAGGTGTAGACGACGACGAACAGAG* |
| p15A-tkDTA-ampR 5' | TTAATAAGATGATCTTCTTGAGATCG |
| p15A-tkDTA-ampR 3' | TTACCAATGCTTAATCAGTGAGG |
| PGK-BSD 5' | TGACTACACCAAGGTGGAAGACAGAGAAAT |
| PGK-BSD 3' | TCGTCACAATAATCTTCCACCGATCGCTTC |
*it is possible to shorten these oligos (however, do not shorten the amplified homology arms for the stop cassette)
For further instructions concerning PCR conditions and the subsequent recombineering pipeline, please see our book chapters in Methods in Enzymology.
For plasmid requests, see here.
The following publications have been cited in our Enzymology Book Chapter about the Gene Targeting Pipeline. For further publications related to Gene Targeting, use of Site Specific Recombinases or Recombineering, please see our publications.
Anastassiadis, K., Fu, J., Patsch, C., Hu, S., Weidlich, S., Duerschke, K., Buchholz, F., Edenhofer, F. and Stewart, A. F. (2009). "Dre recombinase, like Cre, is a highly efficient site-specific recombinase in E. coli, mammalian cells and mice." Dis Model Mech 2: 508-15.
Branda, C. S. and Dymecki, S. M. (2004). "Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice." Dev Cell 6: 7-28.
Buchholz, F., Angrand, P. O. and Stewart, A. F. (1996). "A simple assay to determine the functionality of Cre or FLP recombination targets in genomic manipulation constructs." Nucleic Acids Res 24: 3118-9.
Chen, W. V. and Soriano, P. (2003). "Gene trap mutagenesis in embryonic stem cells." Methods Enzymol 365: 367-86.
Copeland, N. G., Jenkins, N. A. and Court, D. L. (2001). "Recombineering: a powerful new tool for mouse functional genomics." Nat Rev Genet 2: 769-79.
Court, D. L., Sawitzke, J. A. and Thomason, L. C. (2002). "Genetic engineering using homologous recombination." Annu Rev Genet 36: 361-88.
Datsenko, K. A. and Wanner, B. L. (2000). "One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products." Proc Natl Acad Sci U S A 97: 6640-5.
Ellis, H. M., Yu, D., DiTizio, T. and Court, D. L. (2001). "High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides." Proc Natl Acad Sci U S A 98: 6742-6.
Filutowicz, M., McEachern, M. J. and Helinski, D. R. (1986). "Positive and negative roles of an initiator protein at an origin of replication." Proc Natl Acad Sci U S A 83: 9645-9.
Glaser, S., Anastassiadis, K. and Stewart, A. F. (2005). "Current issues in mouse genome engineering." Nat Genet 37: 1187-93.
Hamilton, C. M., Aldea, M., Washburn, B. K., Babitzke, P. and Kushner, S. R. (1989). "New method for generating deletions and gene replacements in Escherichia coli." J Bacteriol 171: 4617-22.
Hashimoto, T. and Sekiguchi, M. (1976). "Isolation of temperature-sensitive mutants of R plasmid by in vitro mutagenesis with hydroxylamine." J Bacteriol 127: 1561-3.
Ivics, Z., Li, M. A., Mates, L., Boeke, J. D., Nagy, A., Bradley, A. and Izsvak, Z. (2009). "Transposon-mediated genome manipulation in vertebrates." Nat Methods 6: 415-22.
Mansour, S. L., Thomas, K. R. and Capecchi, M. R. (1988). "Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes." Nature 336: 348-52.
Poser, I., Sarov, M., Hutchins, J. R., Heriche, J. K., Toyoda, Y., Pozniakovsky, A., Weigl, D., Nitzsche, A., Hegemann, B., Bird, A. W., Pelletier, L., Kittler, R., Hua, S., Naumann, R., Augsburg, M., Sykora, M. M., Hofemeister, H., Zhang, Y., Nasmyth, K., White, K. P., Dietzel, S., Mechtler, K., Durbin, R., Stewart, A. F., Peters, J. M., Buchholz, F. and Hyman, A. A. (2008). "BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals." Nat Methods 5: 409-15.
Ringrose, L., Chabanis, S., Angrand, P. O., Woodroofe, C. and Stewart, A. F. (1999). "Quantitative comparison of DNA looping in vitro and in vivo: chromatin increases effective DNA flexibility at short distances." EMBO J 18: 6630-41.
Sarov, M., Schneider, S., Pozniakovski, A., Roguev, A., Ernst, S., Zhang, Y., Hyman, A. A. and Stewart, A. F. (2006). "A recombineering pipeline for functional genomics applied to Caenorhabditis elegans." Nat Methods 3: 839-44.
Sauer, B. and McDermott, J. (2004). "DNA recombination with a heterospecific Cre homolog identified from comparison of the pac-c1 regions of P1-related phages." Nucleic Acids Res 32: 6086-95.
Sawitzke, J. A., Thomason, L. C., Costantino, N., Bubunenko, M., Datta, S. and Court, D. L. (2007). "Recombineering: in vivo genetic engineering in E. coli, S. enterica, and beyond." Methods Enzymol 421: 171-99.
Skarnes, W., Rosen, B., West, A., Koutsourakis, M., Bushell, W., Iyer, V., Cox, T., Jackson, D., Severin, J., Biggs, P., Thomas, M., Mujica, A., Harrow, J., Fu, J., Nefedov, M., de Jong, P., Stewart, A. and Bradley, A. (2010). "A conditional knockout resource for genome-wide analysis of mouse gene function." Nature in press.
Szymczak, A. L., Workman, C. J., Wang, Y., Vignali, K. M., Dilioglou, S., Vanin, E. F. and Vignali, D. A. (2004). "Correction of multi-gene deficiency in vivo using a single 'self-cleaving' 2A peptide-based retroviral vector." Nat Biotechnol 22: 589-94.
Testa, G., Schaft, J., van der Hoeven, F., Glaser, S., Anastassiadis, K., Zhang, Y., Hermann, T., Stremmel, W. and Stewart, A. F. (2004). "A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles." Genesis 38: 151-8.
Testa, G., Zhang, Y., Vintersten, K., Benes, V., Pijnappel, W. W., Chambers, I., Smith, A. J., Smith, A. G. and Stewart, A. F. (2003). "Engineering the mouse genome with bacterial artificial chromosomes to create multipurpose alleles." Nat Biotechnol 21: 443-7.
Valenzuela, D. M., Murphy, A. J., Frendewey, D., Gale, N. W., Economides, A. N., Auerbach, W., Poueymirou, W. T., Adams, N. C., Rojas, J., Yasenchak, J., Chernomorsky, R., Boucher, M., Elsasser, A. L., Esau, L., Zheng, J., Griffiths, J. A., Wang, X., Su, H., Xue, Y., Dominguez, M. G., Noguera, I., Torres, R., Macdonald, L. E., Stewart, A. F., DeChiara, T. M. and Yancopoulos, G. D. (2003). "High-throughput engineering of the mouse genome coupled with high-resolution expression analysis." Nat Biotechnol 21: 652-9.
Wang, J., Sarov, M., Rientjes, J., Fu, J., Hollak, H., Kranz, H., Xie, W., Stewart, A. F. and Zhang, Y. (2006). "An improved recombineering approach by adding RecA to lambda Red recombination." Mol Biotechnol 32: 43-53.
Wu, S., Wu, Y. and Capecchi, M. R. (2006). "Motoneurons and oligodendrocytes are sequentially generated from neural stem cells but do not appear to share common lineage-restricted progenitors in vivo." Development 133: 581-90.
Wu, S., Ying, G., Wu, Q. and Capecchi, M. R. (2008). "A protocol for constructing gene targeting vectors: generating knockout mice for the cadherin family and beyond." Nat Protoc 3: 1056-76.
Yagi, T., Ikawa, Y., Yoshida, K., Shigetani, Y., Takeda, N., Mabuchi, I., Yamamoto, T. and Aizawa, S. (1990). "Homologous recombination at c-fyn locus of mouse embryonic stem cells with use of diphtheria toxin A-fragment gene in negative selection." Proc Natl Acad Sci U S A 87: 9918-22.
Yang, X. W., Model, P. and Heintz, N. (1997). "Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome." Nat Biotechnol 15: 859-65.
Yang, Y. and Seed, B. (2003). "Site-specific gene targeting in mouse embryonic stem cells with intact bacterial artificial chromosomes." Nat Biotechnol 21: 447-51.
Yu, D., Ellis, H. M., Lee, E. C., Jenkins, N. A., Copeland, N. G. and Court, D. L. (2000). "An efficient recombination system for chromosome engineering in Escherichia coli." Proc Natl Acad Sci U S A 97: 5978-83.
Zhang, Y., Buchholz, F., Muyrers, J. P. and Stewart, A. F. (1998). "A new logic for DNA engineering using recombination in Escherichia coli." Nat Genet 20: 123-8.
Zhang, Y., Muyrers, J. P., Testa, G. and Stewart, A. F. (2000). "DNA cloning by homologous recombination in Escherichia coli." Nat Biotechnol 18: 1314-7.
AMP Ampicillin
AOS Array Oligo Selector
ATG Codon for the initiating methionine
ATT Gateway attachement site (attB, attP, attR, attL)
BAC Bacterial Artificial Chromosome
bb Backbone of a plasmid (origin of replication + selectable marker)
BLAT BLAST Like Alignment Tool
BLAST Basic Local Alignment Search Tool
BSD Blasticidin
CCDS Consensus CDS
CDS Coding sequence for rotein
CE Critical Exon
CHORI Children?s Hospital Oakland Research Institute
CM Chloramphenicol
CNV Copy Number Variant
CoTC Co-Translational Cleavage signal
Cre Site-specific recombinase from bacteriophage P1 genome
CSD CHORI, Sanger Institute, and UC Davis
csm Counter selection marker
D Downstream (oligo)
DAS Distributed Annotation System
Dre Phage D6 derived site-specific DNA recombinase
EGFP Enhanced Green Fluorescent Protein
EJC Exon Junction Complex
EBI European Bioinformatics Institute
EMBL European Molecular Biology Laboratory
EMMA European Mouse Mutant Archive
ENCODE Encyclopedia of DNA Elements
EST Expressed Sequence Tag
EuCOMM European Conditional Mouse Mutagenesis Program
Flp Site-specific recombinase from S.cerevisiae
Flpe Flp, enhanced efficiency at 37 C =
Flpo mammalian codon optimized, enhanced version of Flp
FRT Flp recombinase recognition target
G Gap repair (oligo)
Gen Gentamycin
HA Homology arm
HGNC HUGO Gene Nomenclature Committee
Hprt Hypoxantin phosphoribosyltransferase
HR Homologous Recombination
HRP Horseradish Peroxidase
HUGO Human Genome Organisation
IGTC International Gene Trap Consortium
I-DCC International Data Coordination Centre
IKMC International Knock Out Mouse Consortium
IMMC International Mouse Mutagenesis Consortium
IMSR International Mouse Strain Resource
iPS cells Induced pluripotent stem cells
IRES Internal Ribosome Entry Site
KAN / KM Kanamycin
KO Knock Out
KOMP Knock Out Mouse Project (USA)
loxP Locus of crossover (x) in P1 bacteriophage
LR Longe Range
miRKO micro RNA Knock Out
MGH Massachusetts General Hospital
MGI Mouse Genome Informatics
MGNC Mouse Genome Nomeclature Committee
MGP Mouse Genetics Program
MM(R)RC Mutant Mouse (Regional) Resource Centers
MRC Medical Resarch Council
NCBI National Center for Biotechnology Information
neo Neomycin/Kanamycin
NIH National Institute of Health
NLS Nuclear Localization Signal
NMD Nonsense-Mediated mRNA Decay
NorKOMM North American Conditional Mouse Mutagenesis project (Canada)
OMIM Online Mendelian Inheritance in Man (database)
ori Origin of replication
pA Polyadenylation signal
PB PiggyBAC (Transposon)
PGK promoter Phosphoglycerate kinase promoter
PTC Premature Termination Codon
QC Quality Control
RE Restriction Enzyme site
RMCE Recombinase-Mediaed Cassette Exchange
RMGR Recombinase-Mediaed Genomic Replacement
rox Dre specific recognition site
RRS Recombinase Recognition Site
SB SleepingBeauty (Transposon)
SBP Streptavidin Binding Protein/Peptide
sm Selectable marker
SSR Site Specific Recombinases
Strep Streptomycin
TAP-tag Tandem Affinity Purification
TC Targeting construct
TET Tetracycline
TIGM Texas Institute for Genomic Medicine
U Upstream (oligo)
UCSC University of California Santa Cruz
UTR Untranslated region
VEGA Vertebrate Genome Annotation
WTAC Wellcome Trust Advanced Courses
WTSI Wellcome Trust Sanger Institute
Please contact us for any problems (content or function) of the Gene Targeting Guide.
The author also disclaims accountability for the correctness of the given information and for the content of any external links.
This guide was assembled by Madeleine Walker who thanks all the lab members helping her in gathering the information, especially Jun Fu.