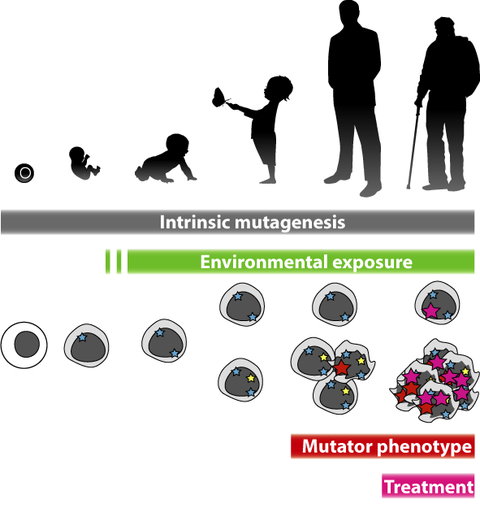
Learning mutagenesis
Mutagenesis is a complicated process to study, because it never occurs with one mechanism in isolation. Both in experimental settings and in every cell of our body, multiple mechanisms are happening simultaneously. Among those mechanisms are stochastic processes, mutagenesis as a result of metabolism, external toxins and radiation, as well as genetic alterations that increase mutagenesis, e.g. by interfering with DNA repair.
Each mutagenic mechanism has different preferences as to where in the genome mutations happen more or less frequently, dependent on sequence preference, DNA repair pathways, and other factors that affect genome specificity of mutagenesis, linked to tissue specific differences, epigenetic modifiers, and co-occurence of multiple mutagenic stimuli simultaneously.
To address this complex interaction of mechanisms in somatic mutagenesis, we are training cascade machine learning models that train on thousands of whole genome sequenced cancer genomes. The models combine classification and regression to encapsulate genome specificity of mutagenesis, usable as an in silico experimental system through hypothesis driven simulations.
In a complementary approach we address per-nucleotide mutation probability through recurrent neural networks on somatic mutagenesis data from individual patients.